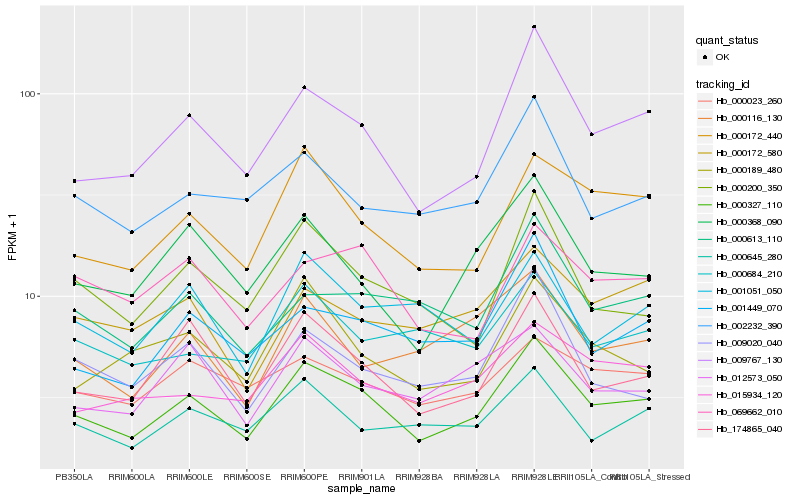
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 2 |
Hb_009767_130 |
0.0773767771 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 3 |
Hb_174865_040 |
0.0973530266 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000172_440 |
0.0986781082 |
- |
- |
PREDICTED: NADPH-dependent 1-acyldihydroxyacetone phosphate reductase [Jatropha curcas] |
| 5 |
Hb_000368_090 |
0.1058933291 |
- |
- |
TPA: hypothetical protein ZEAMMB73_442906 [Zea mays] |
| 6 |
Hb_001449_070 |
0.1076787376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_015934_120 |
0.1081766435 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |
| 8 |
Hb_002232_390 |
0.1095269975 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 9 |
Hb_000645_280 |
0.1113547699 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 10 |
Hb_000200_350 |
0.1114848385 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 11 |
Hb_000613_110 |
0.1143674073 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 12 |
Hb_000189_480 |
0.1164835976 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 13 |
Hb_069662_010 |
0.1165699428 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 14 |
Hb_012573_050 |
0.1176411922 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 15 |
Hb_000023_260 |
0.1190449016 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_009020_040 |
0.1191670551 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000684_210 |
0.1195114665 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 18 |
Hb_001051_050 |
0.1201753253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000116_130 |
0.1211826041 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 20 |
Hb_000172_580 |
0.1218650776 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |