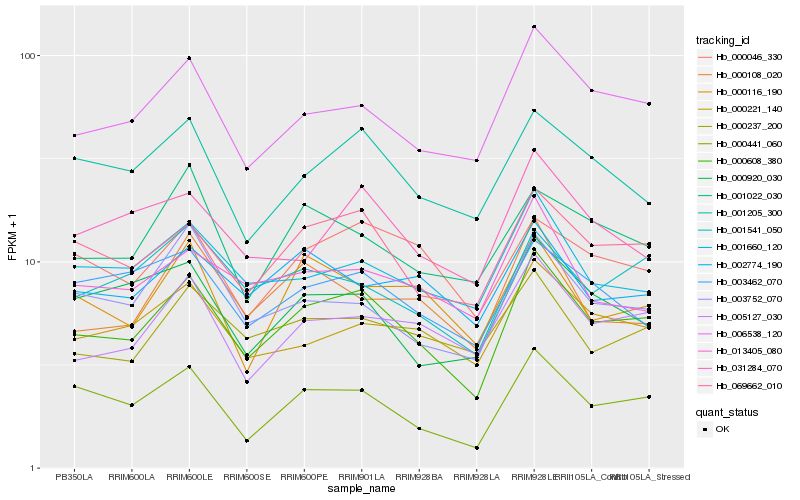
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_380 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_069662_010 |
0.0821192795 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 3 |
Hb_006538_120 |
0.0883989219 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000441_060 |
0.0927088905 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 5 |
Hb_005127_030 |
0.1007102804 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 6 |
Hb_000221_140 |
0.1067239523 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_001205_300 |
0.1084809866 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 8 |
Hb_001660_120 |
0.1094525284 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000237_200 |
0.109549857 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 10 |
Hb_000046_330 |
0.111117027 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |
| 11 |
Hb_001541_050 |
0.1130260589 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 12 |
Hb_003752_070 |
0.1142770318 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 13 |
Hb_031284_070 |
0.1159016303 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000920_030 |
0.1166466232 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_013405_080 |
0.1166575149 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 16 |
Hb_000108_020 |
0.1168128972 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 17 |
Hb_000116_190 |
0.1179707669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002774_190 |
0.1180153668 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 19 |
Hb_003462_070 |
0.1184726082 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 20 |
Hb_001022_030 |
0.1185868023 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |