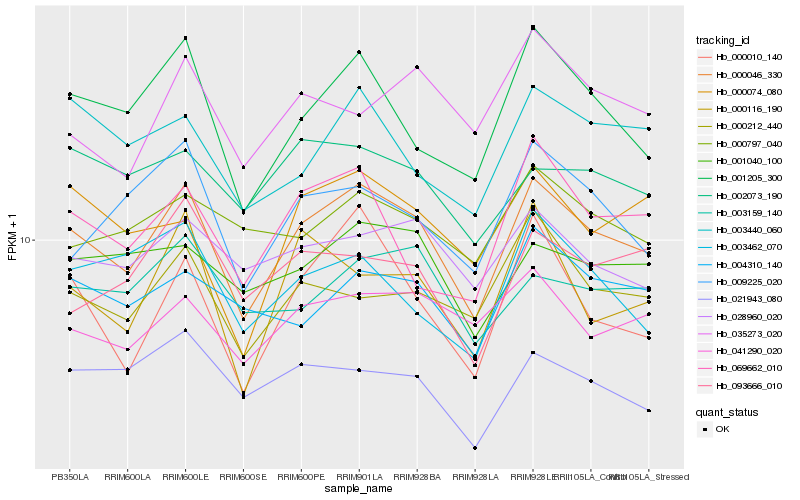
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_330 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |
| 2 |
Hb_001205_300 |
0.0868595779 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 3 |
Hb_028960_020 |
0.0982964357 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000212_440 |
0.0987513841 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_041290_020 |
0.099686392 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 6 |
Hb_001040_100 |
0.0997274808 |
- |
- |
PREDICTED: general transcription factor IIH subunit 4 [Jatropha curcas] |
| 7 |
Hb_000010_140 |
0.1007388894 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 11, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_069662_010 |
0.1012887525 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 9 |
Hb_021943_080 |
0.1016262739 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009225_020 |
0.1017233438 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 11 |
Hb_004310_140 |
0.1030316648 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_003440_060 |
0.1035963064 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 13 |
Hb_003159_140 |
0.1039443044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000074_080 |
0.104831285 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_002073_190 |
0.1063399431 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003462_070 |
0.1075709436 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 17 |
Hb_000797_040 |
0.1079596694 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 18 |
Hb_035273_020 |
0.1080239924 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_093666_010 |
0.1082920858 |
- |
- |
BnaC05g06620D [Brassica napus] |
| 20 |
Hb_000116_190 |
0.1088520643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |