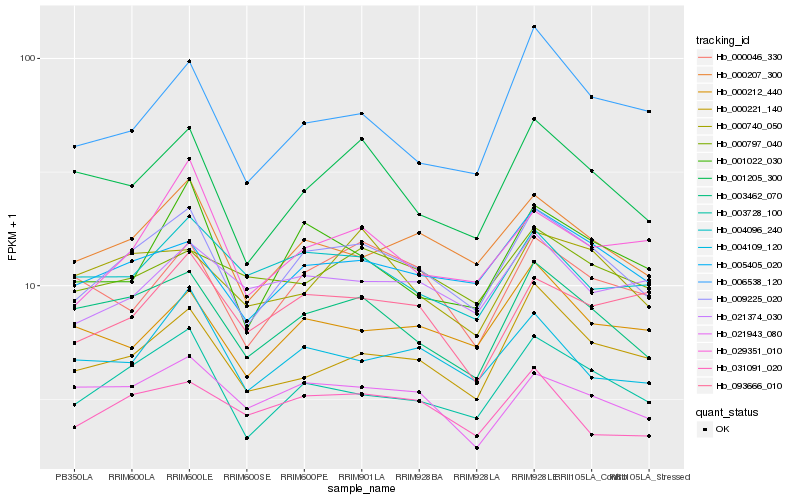
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009225_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 2 |
Hb_003728_100 |
0.0623837965 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000207_300 |
0.072353124 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_005405_020 |
0.0758978373 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_003462_070 |
0.0770397768 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 6 |
Hb_001205_300 |
0.0876609167 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 7 |
Hb_000221_140 |
0.0902492099 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001022_030 |
0.0946787722 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 9 |
Hb_004109_120 |
0.0991232145 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000212_440 |
0.1006225095 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000046_330 |
0.1017233438 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |
| 12 |
Hb_029351_010 |
0.1024321308 |
- |
- |
PREDICTED: HD domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_021943_080 |
0.1028092558 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_093666_010 |
0.103335131 |
- |
- |
BnaC05g06620D [Brassica napus] |
| 15 |
Hb_000740_050 |
0.103605684 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 16 |
Hb_006538_120 |
0.1038188725 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 17 |
Hb_004096_240 |
0.1042487046 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 18 |
Hb_031091_020 |
0.1050342054 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 19 |
Hb_021374_030 |
0.1060160393 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 20 |
Hb_000797_040 |
0.1064178697 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |