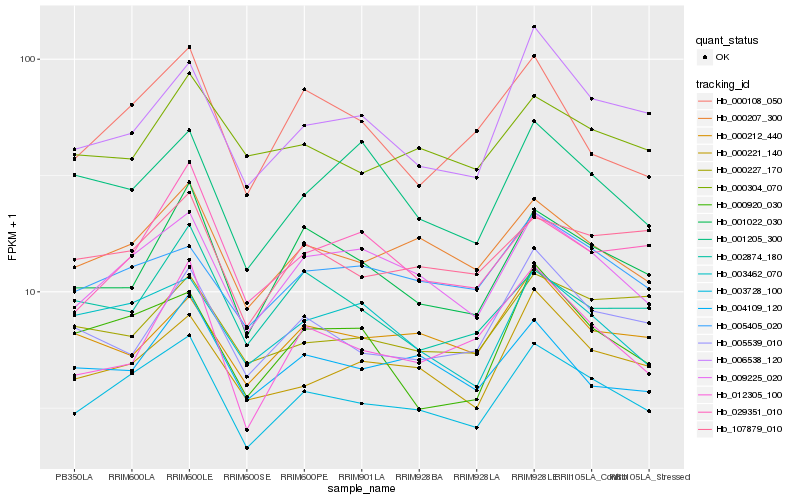
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003728_100 |
0.0 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009225_020 |
0.0623837965 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 3 |
Hb_000207_300 |
0.068257033 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000221_140 |
0.0866007668 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_003462_070 |
0.0899330017 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 6 |
Hb_001022_030 |
0.0907589592 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 7 |
Hb_005405_020 |
0.0918338708 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 8 |
Hb_107879_010 |
0.0918966853 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 9 |
Hb_006538_120 |
0.0952691788 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000920_030 |
0.1001655285 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_004109_120 |
0.1006478607 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000304_070 |
0.1032849146 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 13 |
Hb_005539_010 |
0.1036258752 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 14 |
Hb_000108_050 |
0.1045073463 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 15 |
Hb_029351_010 |
0.1045157851 |
- |
- |
PREDICTED: HD domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_012305_100 |
0.1056738314 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000212_440 |
0.1062267855 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_002874_180 |
0.1078155832 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001205_300 |
0.1081672419 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 20 |
Hb_000227_170 |
0.1107054265 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |