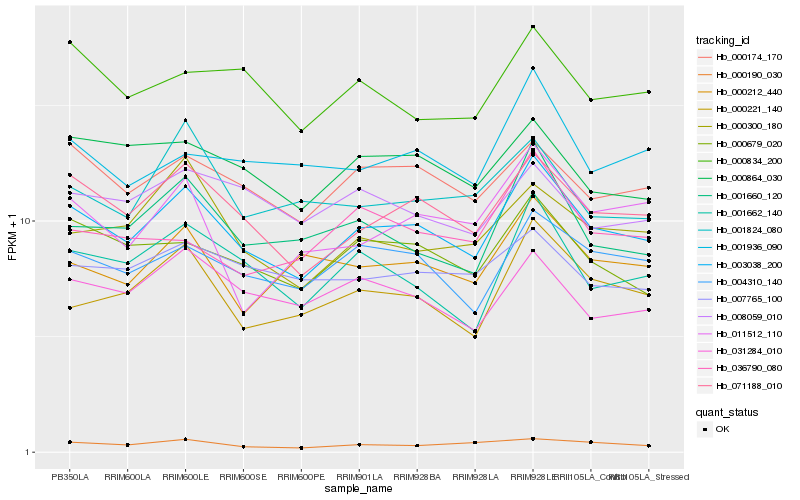
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003038_200 |
0.0 |
desease resistance |
Gene Name: MMR_HSR1 |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |
| 2 |
Hb_071188_010 |
0.0557162199 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 3 |
Hb_004310_140 |
0.0760818643 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_000679_020 |
0.0807990606 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 5 |
Hb_000221_140 |
0.082131872 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000174_170 |
0.0834011503 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001662_140 |
0.0834974123 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001660_120 |
0.0860461911 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000190_030 |
0.0880818054 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA-like [Pyrus x bretschneideri] |
| 10 |
Hb_031284_010 |
0.0912918879 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 11 |
Hb_000864_030 |
0.092249877 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000212_440 |
0.0934533319 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_000834_200 |
0.09561303 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 14 |
Hb_001936_090 |
0.0958496567 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000300_180 |
0.0967693487 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 16 |
Hb_007765_100 |
0.0988829898 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 17 |
Hb_008059_010 |
0.1018483275 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001824_080 |
0.1020785738 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 19 |
Hb_036790_080 |
0.1021920994 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 20 |
Hb_011512_110 |
0.1025776068 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |