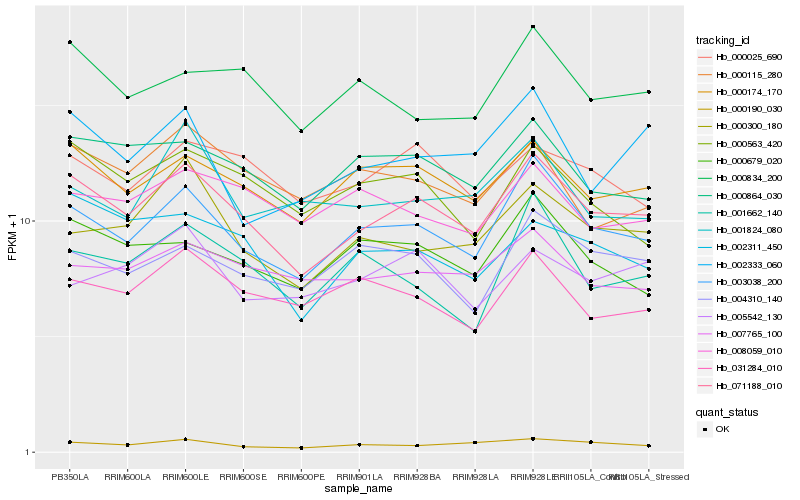
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_071188_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 2 |
Hb_003038_200 |
0.0557162199 |
desease resistance |
Gene Name: MMR_HSR1 |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000174_170 |
0.073546663 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000864_030 |
0.085297355 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000190_030 |
0.0910157959 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA-like [Pyrus x bretschneideri] |
| 6 |
Hb_002311_450 |
0.0933788627 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 7 |
Hb_000025_690 |
0.0938719433 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 8 |
Hb_000679_020 |
0.0938984991 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 9 |
Hb_000300_180 |
0.0941687893 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 10 |
Hb_000834_200 |
0.0960131074 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 11 |
Hb_007765_100 |
0.0963874973 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 12 |
Hb_004310_140 |
0.0979759638 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_000563_420 |
0.0999992979 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 14 |
Hb_031284_010 |
0.1000506851 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 15 |
Hb_000115_280 |
0.1014945043 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 16 |
Hb_002333_060 |
0.1032424454 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_008059_010 |
0.1035969923 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001662_140 |
0.1074825773 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005542_130 |
0.1096159284 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 20 |
Hb_001824_080 |
0.1099759233 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |