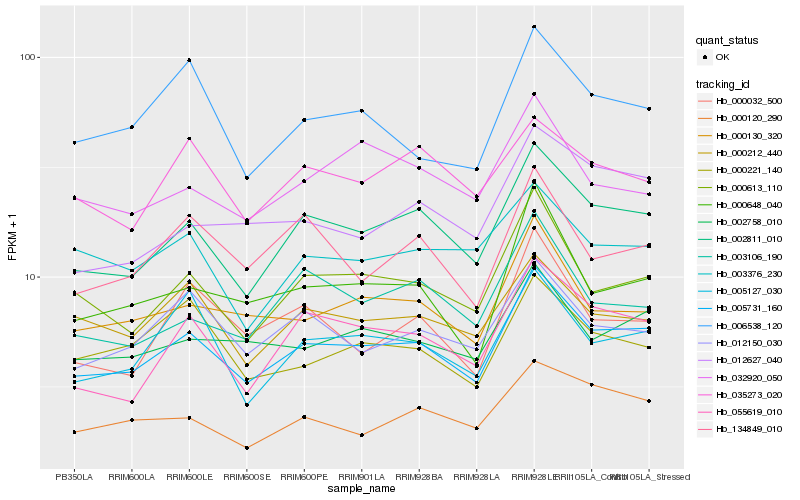
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005731_160 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_002811_010 |
0.0468953117 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 3 |
Hb_000613_110 |
0.0703583266 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 4 |
Hb_002758_010 |
0.078509611 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 5 |
Hb_000130_320 |
0.0792488711 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 6 |
Hb_055619_010 |
0.0806420038 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000648_040 |
0.0839370293 |
- |
- |
unknown [Populus trichocarpa] |
| 8 |
Hb_012150_030 |
0.0858641035 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000120_290 |
0.0867928542 |
- |
- |
PREDICTED: N-acylphosphatidylethanolamine synthase isoform X1 [Jatropha curcas] |
| 10 |
Hb_005127_030 |
0.0871429257 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 11 |
Hb_000212_440 |
0.0908563031 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_006538_120 |
0.0923614344 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003106_190 |
0.0925517505 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 14 |
Hb_012627_040 |
0.0932635567 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 15 |
Hb_134849_010 |
0.0937171877 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 16 |
Hb_000221_140 |
0.0948443818 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_032920_050 |
0.0968194442 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000032_500 |
0.0973362857 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 19 |
Hb_035273_020 |
0.0998991168 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_003376_230 |
0.0999324173 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |