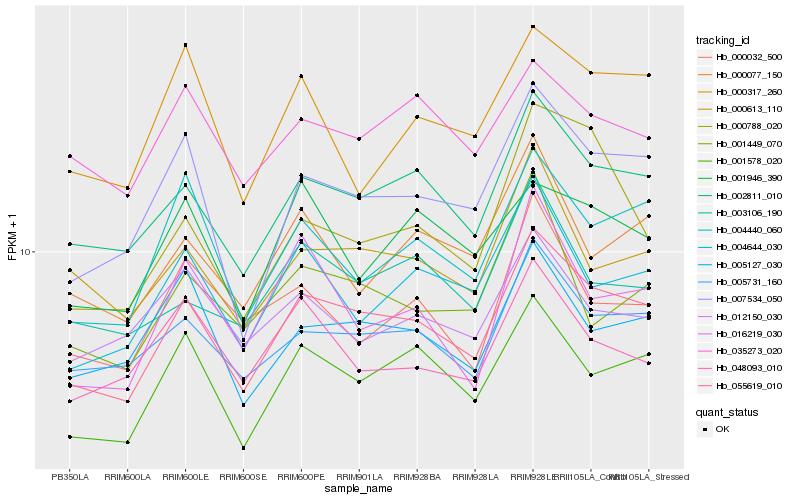
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055619_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002811_010 |
0.0624700854 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 3 |
Hb_005731_160 |
0.0806420038 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_007534_050 |
0.0813867607 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_001578_020 |
0.0842114509 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 6 |
Hb_004440_060 |
0.0964655031 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 7 |
Hb_004644_030 |
0.0984679697 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 8 |
Hb_005127_030 |
0.1019749496 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_000613_110 |
0.102647259 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 10 |
Hb_003106_190 |
0.1078794321 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 11 |
Hb_000077_150 |
0.1085483795 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 12 |
Hb_035273_020 |
0.1130186137 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001946_390 |
0.1151189187 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 14 |
Hb_016219_030 |
0.1152089359 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_012150_030 |
0.1156145323 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001449_070 |
0.1158981247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_048093_010 |
0.1166092583 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_000788_020 |
0.1166181508 |
- |
- |
PREDICTED: uncharacterized protein LOC105629709 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000317_260 |
0.1169829194 |
- |
- |
unknown [Populus trichocarpa] |
| 20 |
Hb_000032_500 |
0.1170242623 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |