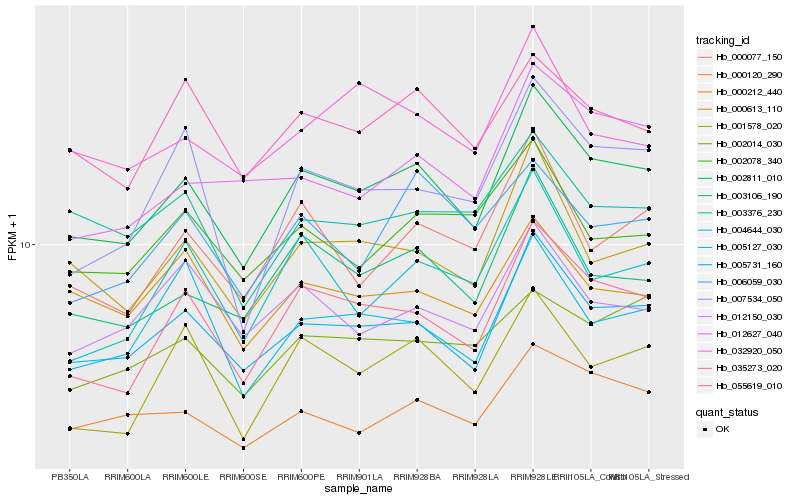
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_010 |
0.0 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 2 |
Hb_005731_160 |
0.0468953117 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_055619_010 |
0.0624700854 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003106_190 |
0.0761116356 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 5 |
Hb_000613_110 |
0.0780668772 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 6 |
Hb_000120_290 |
0.0836677816 |
- |
- |
PREDICTED: N-acylphosphatidylethanolamine synthase isoform X1 [Jatropha curcas] |
| 7 |
Hb_000077_150 |
0.0876176722 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 8 |
Hb_035273_020 |
0.090049858 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001578_020 |
0.0963556742 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 10 |
Hb_004644_030 |
0.0965362702 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 11 |
Hb_002078_340 |
0.0978610669 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 12 |
Hb_012627_040 |
0.0978795466 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 13 |
Hb_012150_030 |
0.0987723522 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_007534_050 |
0.098772444 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000212_440 |
0.1000900819 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_003376_230 |
0.100283047 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 17 |
Hb_005127_030 |
0.1007368799 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 18 |
Hb_002014_030 |
0.1017119254 |
- |
- |
hypothetical protein JCGZ_07251 [Jatropha curcas] |
| 19 |
Hb_032920_050 |
0.1023193229 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_006059_030 |
0.1032037556 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |