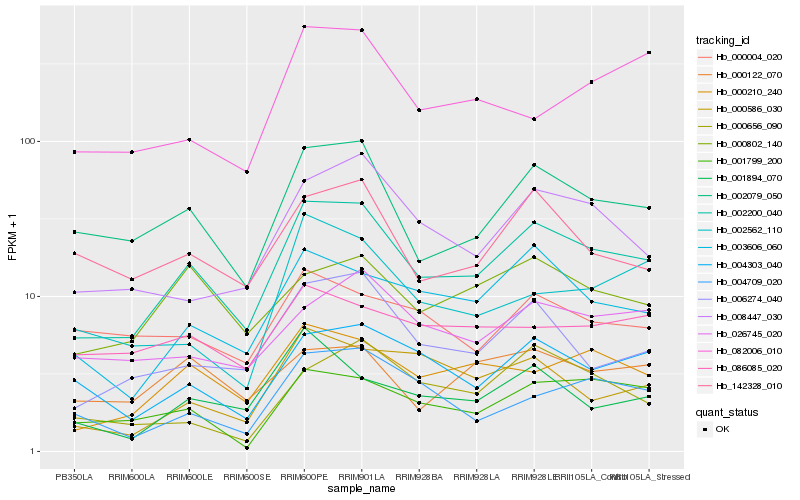
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002200_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002079_050 |
0.1106406315 |
- |
- |
hypothetical protein B456_004G182300 [Gossypium raimondii] |
| 3 |
Hb_000210_240 |
0.1273203413 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 4 |
Hb_006274_040 |
0.1280781513 |
- |
- |
PREDICTED: DENN domain and WD repeat-containing protein SCD1-like [Fragaria vesca subsp. vesca] |
| 5 |
Hb_003606_060 |
0.1288737825 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 6 |
Hb_004303_040 |
0.1382224322 |
- |
- |
PREDICTED: fumarate hydratase 1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_008447_030 |
0.1417683882 |
- |
- |
- |
| 8 |
Hb_001799_200 |
0.1442884677 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 9 |
Hb_142328_010 |
0.1475737977 |
- |
- |
PREDICTED: uncharacterized protein LOC105630513 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000802_140 |
0.1484013166 |
- |
- |
hypothetical protein B456_011G045700 [Gossypium raimondii] |
| 11 |
Hb_026745_020 |
0.1485924839 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 8, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004709_020 |
0.1575367587 |
- |
- |
PREDICTED: putative threonine aspartase [Jatropha curcas] |
| 13 |
Hb_000122_070 |
0.1591089776 |
- |
- |
hypothetical protein POPTR_0010s18960g [Populus trichocarpa] |
| 14 |
Hb_000586_030 |
0.1603537026 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 15 |
Hb_001894_070 |
0.1624321192 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_086085_020 |
0.1653544379 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 17 |
Hb_002562_110 |
0.1684038779 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000004_020 |
0.1688388313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_082006_010 |
0.1713830472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000656_090 |
0.1716847125 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |