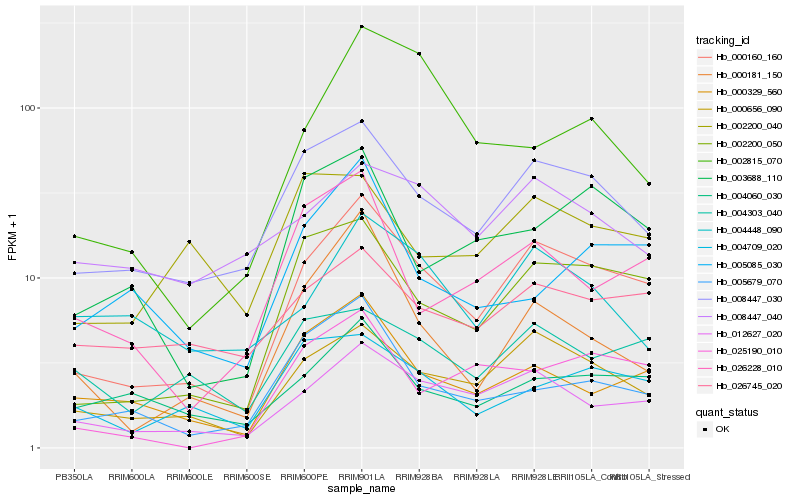
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000160_160 |
0.0 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002200_050 |
0.1225682078 |
- |
- |
unknown [Lotus japonicus] |
| 3 |
Hb_000656_090 |
0.1273816639 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 4 |
Hb_012627_020 |
0.1274099168 |
- |
- |
PHYTOCHROME AND FLOWERING TIME 1 family protein [Populus trichocarpa] |
| 5 |
Hb_008447_030 |
0.1432604126 |
- |
- |
- |
| 6 |
Hb_000329_560 |
0.1752508018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_025190_010 |
0.1837652881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004060_030 |
0.1863889254 |
- |
- |
PREDICTED: primase homolog protein [Jatropha curcas] |
| 9 |
Hb_003688_110 |
0.1876205759 |
- |
- |
- |
| 10 |
Hb_000181_150 |
0.1891518761 |
- |
- |
hypothetical protein PHAVU_011G1647000g, partial [Phaseolus vulgaris] |
| 11 |
Hb_026745_020 |
0.1914877207 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 8, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004709_020 |
0.1949624488 |
- |
- |
PREDICTED: putative threonine aspartase [Jatropha curcas] |
| 13 |
Hb_026228_010 |
0.1952452761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005679_070 |
0.1967270361 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 15 |
Hb_004303_040 |
0.1974303844 |
- |
- |
PREDICTED: fumarate hydratase 1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_005085_030 |
0.2093196945 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_002815_070 |
0.2115055623 |
- |
- |
hypothetical protein PHAVU_005G048300g [Phaseolus vulgaris] |
| 18 |
Hb_004448_090 |
0.2120794968 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 19 |
Hb_008447_040 |
0.213797993 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 20 |
Hb_002200_040 |
0.2158189657 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X1 [Jatropha curcas] |