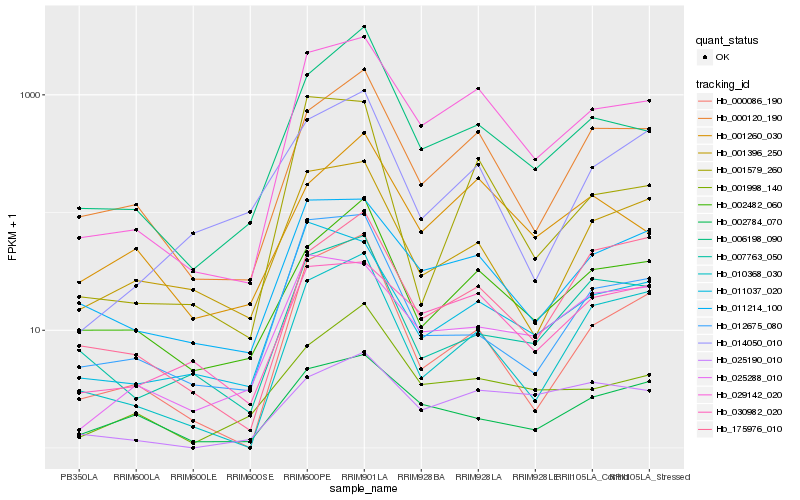
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029142_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s23900g [Populus trichocarpa] |
| 2 |
Hb_000120_190 |
0.1442353212 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 3 |
Hb_011214_100 |
0.1500760166 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 4 |
Hb_006198_090 |
0.15602109 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 5 |
Hb_030982_020 |
0.1583480481 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 6 |
Hb_011037_020 |
0.163535954 |
- |
- |
unknown [Picea sitchensis] |
| 7 |
Hb_010368_030 |
0.1671488581 |
- |
- |
BnaA07g35570D [Brassica napus] |
| 8 |
Hb_000086_190 |
0.1672268408 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 9 |
Hb_001998_140 |
0.1674983555 |
- |
- |
- |
| 10 |
Hb_025288_010 |
0.171958836 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 11 |
Hb_002482_060 |
0.1740515975 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 12 |
Hb_025190_010 |
0.1789498788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001579_260 |
0.1856324175 |
- |
- |
putative ADP-ribolylation factor [Arabidopsis thaliana] |
| 14 |
Hb_001396_250 |
0.1877285188 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 15 |
Hb_001260_030 |
0.188600447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_012675_080 |
0.1911848186 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 17 |
Hb_007763_050 |
0.191263608 |
- |
- |
hypothetical protein L484_012752 [Morus notabilis] |
| 18 |
Hb_014050_010 |
0.1931284905 |
- |
- |
PREDICTED: histone H3.3-like, partial [Phoenix dactylifera] |
| 19 |
Hb_002784_070 |
0.1970609329 |
- |
- |
BnaUnng01470D [Brassica napus] |
| 20 |
Hb_175976_010 |
0.2027059975 |
- |
- |
- |