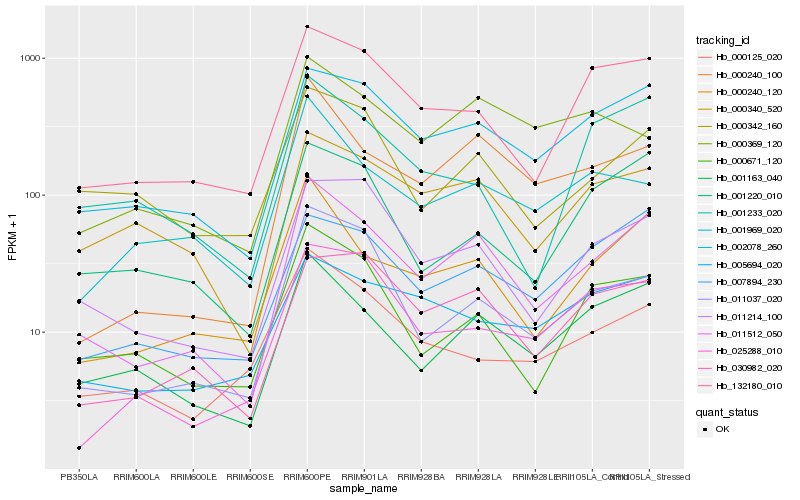
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011512_050 |
0.0 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 2 |
Hb_002078_260 |
0.1190487904 |
- |
- |
unknown [Populus trichocarpa] |
| 3 |
Hb_001163_040 |
0.1208528915 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 4 |
Hb_000240_120 |
0.1233281592 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 5 |
Hb_000240_100 |
0.1369220817 |
- |
- |
PREDICTED: ras-related protein Rab5-like [Eucalyptus grandis] |
| 6 |
Hb_132180_010 |
0.1455832611 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 7 |
Hb_011214_100 |
0.1477036447 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 8 |
Hb_011037_020 |
0.1509604222 |
- |
- |
unknown [Picea sitchensis] |
| 9 |
Hb_007894_230 |
0.1517234304 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 10 |
Hb_030982_020 |
0.1539552111 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_000671_120 |
0.1541425032 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 12 |
Hb_000340_520 |
0.1561174962 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 13 |
Hb_001220_010 |
0.1621536314 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 14 |
Hb_000342_160 |
0.1662700427 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 15 |
Hb_001969_020 |
0.1733667745 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 16 |
Hb_000125_020 |
0.1746239569 |
transcription factor |
TF Family: mTERF |
- |
| 17 |
Hb_005694_020 |
0.1749869648 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 18 |
Hb_025288_010 |
0.1757989987 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 19 |
Hb_001233_020 |
0.1795590902 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 20 |
Hb_000369_120 |
0.1816688752 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3-B [Jatropha curcas] |