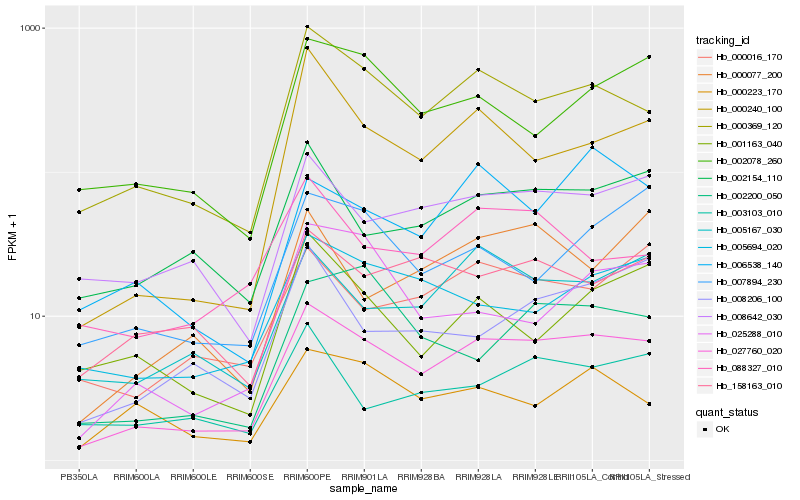
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027760_020 |
0.0 |
- |
- |
hypothetical protein M569_04462, partial [Genlisea aurea] |
| 2 |
Hb_000369_120 |
0.1128911464 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3-B [Jatropha curcas] |
| 3 |
Hb_008642_030 |
0.1515967165 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005167_030 |
0.1535490211 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like, partial [Camelina sativa] |
| 5 |
Hb_002154_110 |
0.1550048969 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 6 |
Hb_000016_170 |
0.1641124508 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005694_020 |
0.1647281582 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 8 |
Hb_002078_260 |
0.1670050222 |
- |
- |
unknown [Populus trichocarpa] |
| 9 |
Hb_158163_010 |
0.168216466 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_000240_100 |
0.1686396833 |
- |
- |
PREDICTED: ras-related protein Rab5-like [Eucalyptus grandis] |
| 11 |
Hb_088327_010 |
0.1791327505 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 12 |
Hb_006538_140 |
0.1808417412 |
- |
- |
PREDICTED: uncharacterized protein LOC103341483 [Prunus mume] |
| 13 |
Hb_000223_170 |
0.1816316715 |
- |
- |
- |
| 14 |
Hb_008206_100 |
0.1828181022 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 15 |
Hb_007894_230 |
0.1829937107 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 16 |
Hb_003103_010 |
0.1831060676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000077_200 |
0.1839055836 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 18 |
Hb_025288_010 |
0.1842281457 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 19 |
Hb_001163_040 |
0.1861757267 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 20 |
Hb_002200_050 |
0.1863693119 |
- |
- |
unknown [Lotus japonicus] |