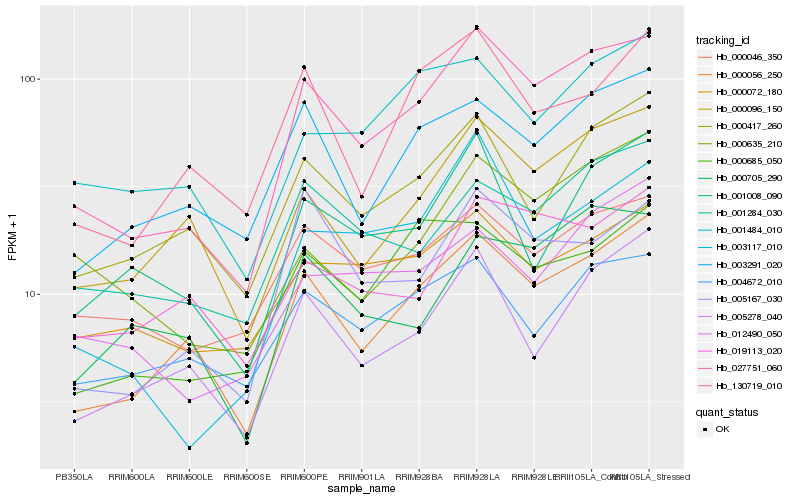
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027751_060 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B [Jatropha curcas] |
| 2 |
Hb_000056_250 |
0.0907172472 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_000096_150 |
0.1134993489 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 4 |
Hb_000046_350 |
0.1158506385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005167_030 |
0.1208251742 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like, partial [Camelina sativa] |
| 6 |
Hb_000417_260 |
0.1240379278 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_001284_030 |
0.1244186868 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_003117_010 |
0.1245871714 |
- |
- |
PREDICTED: uncharacterized protein LOC100259103 isoform X1 [Vitis vinifera] |
| 9 |
Hb_003291_020 |
0.1258438491 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 10 |
Hb_005278_040 |
0.1265751602 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 11 |
Hb_000685_050 |
0.1268152063 |
- |
- |
PREDICTED: mitochondrial inner membrane protein OXA1-like [Populus euphratica] |
| 12 |
Hb_000072_180 |
0.1276823088 |
- |
- |
PREDICTED: EVI5-like protein [Jatropha curcas] |
| 13 |
Hb_001008_090 |
0.129069335 |
- |
- |
cytochrome C oxidase, putative [Ricinus communis] |
| 14 |
Hb_130719_010 |
0.1300661454 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 15 |
Hb_004672_010 |
0.1301608041 |
- |
- |
ecotropic viral integration site, putative [Ricinus communis] |
| 16 |
Hb_001484_010 |
0.1302619057 |
- |
- |
hypothetical protein POPTR_0005s07440g [Populus trichocarpa] |
| 17 |
Hb_000635_210 |
0.1316151575 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_012490_050 |
0.1341735275 |
- |
- |
PREDICTED: uncharacterized protein LOC105637921 [Jatropha curcas] |
| 19 |
Hb_019113_020 |
0.1354233011 |
- |
- |
hypothetical protein JCGZ_13398 [Jatropha curcas] |
| 20 |
Hb_000705_290 |
0.1355888841 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |