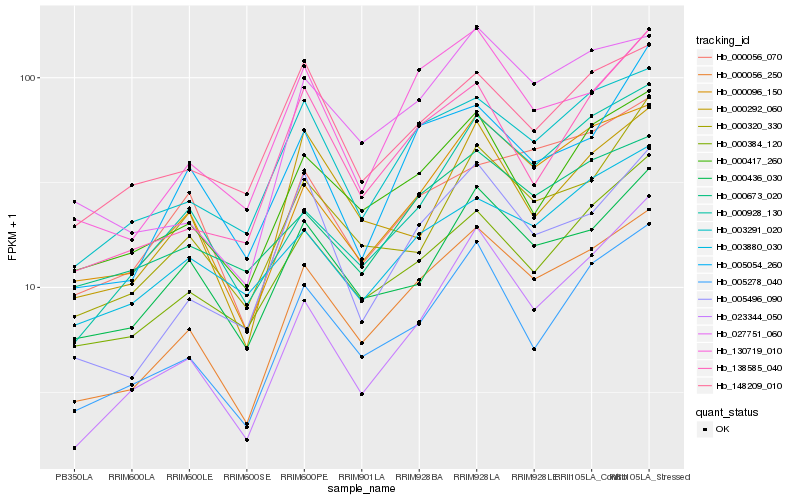
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000056_250 |
0.0 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 2 |
Hb_000096_150 |
0.0739935779 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 3 |
Hb_003291_020 |
0.0888261992 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 4 |
Hb_005278_040 |
0.0899273247 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 5 |
Hb_027751_060 |
0.0907172472 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B [Jatropha curcas] |
| 6 |
Hb_000417_260 |
0.0927043757 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_130719_010 |
0.0943234505 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 8 |
Hb_000436_030 |
0.101571408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005496_090 |
0.1016466683 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 10 |
Hb_000384_120 |
0.1039168406 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 11 |
Hb_005054_260 |
0.1135723745 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 12 |
Hb_000673_020 |
0.1137735627 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 13 |
Hb_000320_330 |
0.1149354964 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |
| 14 |
Hb_000056_070 |
0.1154350175 |
- |
- |
- |
| 15 |
Hb_000928_130 |
0.1160741776 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_023344_050 |
0.1167644257 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 17 |
Hb_148209_010 |
0.1174298815 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 18 |
Hb_003880_030 |
0.1184809558 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 19 |
Hb_138585_040 |
0.1193132621 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 20 |
Hb_000292_060 |
0.1203727563 |
- |
- |
transporter, putative [Ricinus communis] |