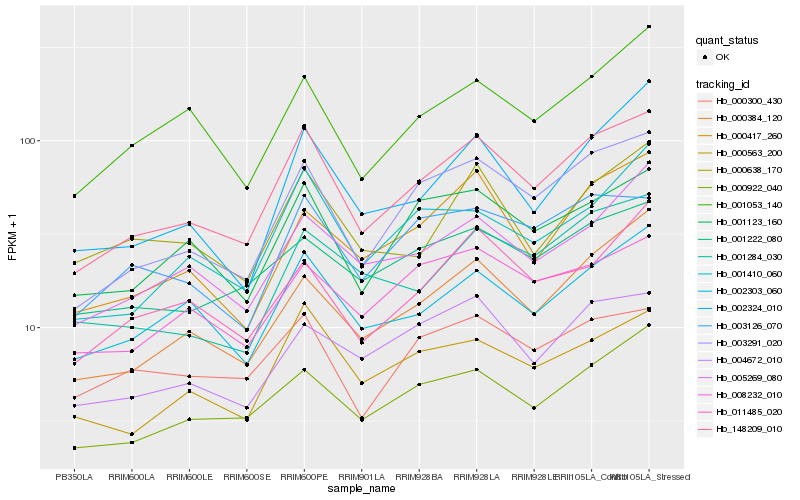
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148209_010 |
0.0 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 2 |
Hb_003291_020 |
0.0550034782 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 3 |
Hb_002303_060 |
0.0824889687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001123_160 |
0.0833286149 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 5 |
Hb_000922_040 |
0.0850570062 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 6 |
Hb_000563_200 |
0.0858694507 |
- |
- |
hypothetical protein JCGZ_18930 [Jatropha curcas] |
| 7 |
Hb_000384_120 |
0.0900299247 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001410_060 |
0.0932800421 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 9 |
Hb_005269_080 |
0.0942325262 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_001053_140 |
0.0946806807 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 11 |
Hb_003126_070 |
0.0949354447 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000417_260 |
0.0969826744 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_004672_010 |
0.0976989293 |
- |
- |
ecotropic viral integration site, putative [Ricinus communis] |
| 14 |
Hb_008232_010 |
0.0995681712 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000300_430 |
0.0999037256 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 16 |
Hb_002324_010 |
0.1003230422 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 17 |
Hb_011485_020 |
0.1005280545 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_001222_080 |
0.1009500606 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000638_170 |
0.1016627892 |
- |
- |
PREDICTED: ras-related protein RABC1 [Jatropha curcas] |
| 20 |
Hb_001284_030 |
0.1020681846 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |