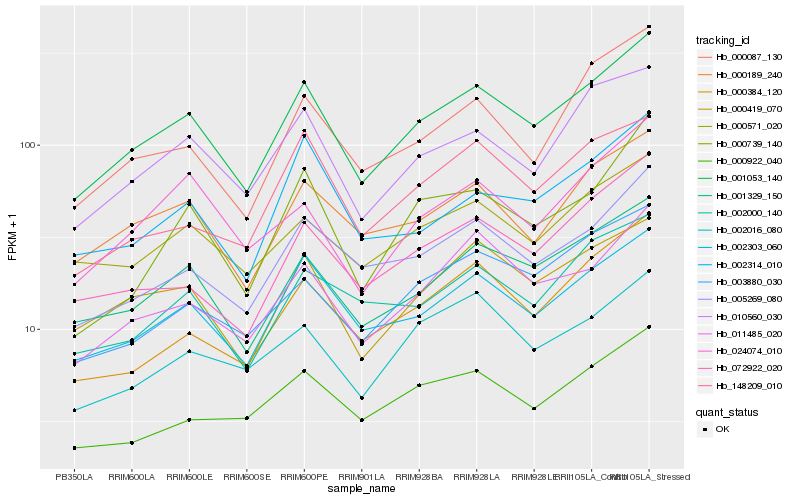
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001053_140 |
0.0 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 2 |
Hb_001329_150 |
0.058890699 |
- |
- |
PREDICTED: autophagy-related protein 8i-like [Jatropha curcas] |
| 3 |
Hb_002303_060 |
0.0603934455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000419_070 |
0.0712312818 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 5 |
Hb_010560_030 |
0.0722552678 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |
| 6 |
Hb_000189_240 |
0.0737805455 |
- |
- |
PREDICTED: PITH domain-containing protein At3g04780 [Jatropha curcas] |
| 7 |
Hb_005269_080 |
0.0746135569 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_011485_020 |
0.0754570565 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000571_020 |
0.0827363544 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_072922_020 |
0.0867770001 |
- |
- |
SNARE-like superfamily protein [Theobroma cacao] |
| 11 |
Hb_003880_030 |
0.0871589309 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002000_140 |
0.0877744343 |
- |
- |
PREDICTED: thioredoxin Y2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000384_120 |
0.0883630905 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 14 |
Hb_024074_010 |
0.0903635484 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 15 |
Hb_000087_130 |
0.0911397711 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial [Jatropha curcas] |
| 16 |
Hb_002016_080 |
0.0920062977 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000922_040 |
0.0942582127 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 18 |
Hb_148209_010 |
0.0946806807 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 19 |
Hb_002314_010 |
0.0947921342 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 20 |
Hb_000739_140 |
0.09480202 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |