| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
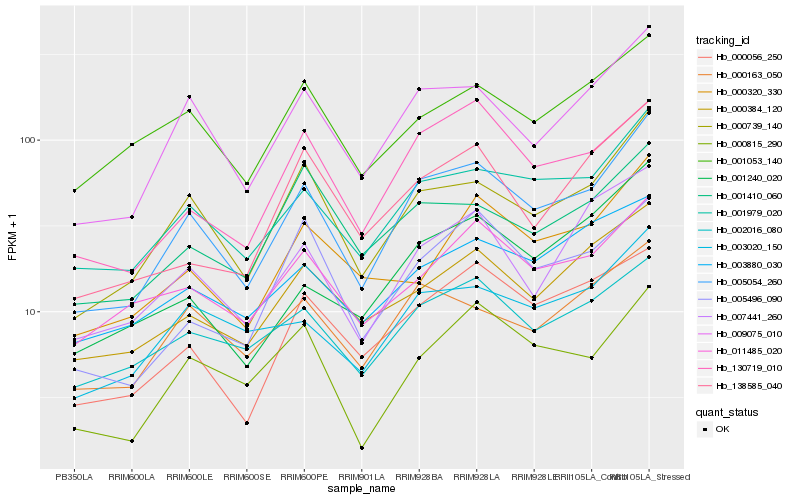
Hb_005054_260 |
0.0 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 2 |
Hb_000739_140 |
0.0752162047 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 3 |
Hb_009075_010 |
0.0767191261 |
- |
- |
60S ribosomal protein L10B [Hevea brasiliensis] |
| 4 |
Hb_001979_020 |
0.080959662 |
- |
- |
hypothetical protein OsJ_31823 [Oryza sativa Japonica Group] |
| 5 |
Hb_003020_150 |
0.1093191982 |
- |
- |
tropinone reductase, putative [Ricinus communis] |
| 6 |
Hb_000384_120 |
0.1098382809 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000163_050 |
0.1099696969 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |
| 8 |
Hb_002016_080 |
0.1128472482 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000056_250 |
0.1135723745 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 10 |
Hb_130719_010 |
0.1138376615 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 11 |
Hb_007441_260 |
0.1139713151 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_011485_020 |
0.1180176415 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000815_290 |
0.1199591689 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 14 |
Hb_005496_090 |
0.1227755266 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_001240_020 |
0.1228079084 |
- |
- |
PREDICTED: uncharacterized protein LOC105641171 [Jatropha curcas] |
| 16 |
Hb_001053_140 |
0.1230029215 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 17 |
Hb_138585_040 |
0.124013265 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 18 |
Hb_003880_030 |
0.1249869801 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001410_060 |
0.1261801372 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 20 |
Hb_000320_330 |
0.1273494072 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |