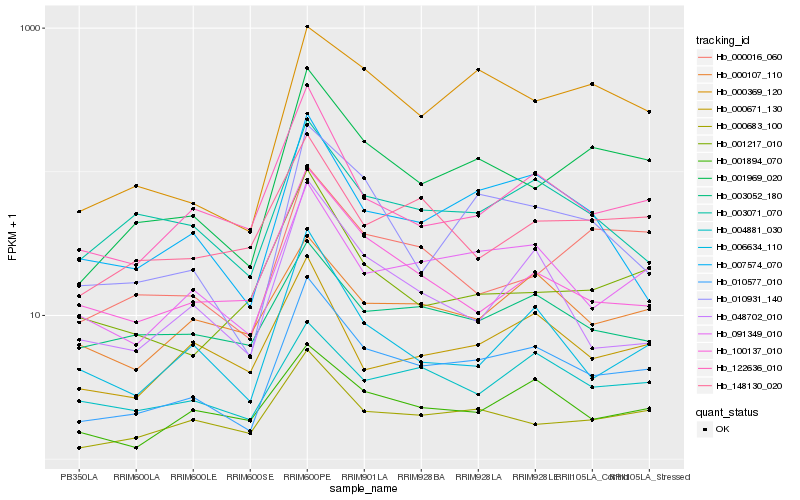
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010577_010 |
0.0 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 2 |
Hb_091349_010 |
0.110933562 |
- |
- |
hypothetical protein EUGRSUZ_C02287 [Eucalyptus grandis] |
| 3 |
Hb_001969_020 |
0.1189499758 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 4 |
Hb_000683_100 |
0.1316859199 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 5 |
Hb_122636_010 |
0.1387633777 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 6 |
Hb_010931_140 |
0.1399524603 |
- |
- |
- |
| 7 |
Hb_007574_070 |
0.146243363 |
- |
- |
fatty acid desaturase, putative [Ricinus communis] |
| 8 |
Hb_006634_110 |
0.1468144073 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 9 |
Hb_001894_070 |
0.147497439 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003071_070 |
0.1499256582 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 11 |
Hb_000369_120 |
0.1520233292 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3-B [Jatropha curcas] |
| 12 |
Hb_000671_130 |
0.1578633709 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_048702_010 |
0.160260349 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_000107_110 |
0.1621769571 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_004881_030 |
0.1623890191 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 16 |
Hb_100137_010 |
0.1651728313 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 17 |
Hb_003052_180 |
0.1655944313 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 18 |
Hb_148130_020 |
0.166642533 |
- |
- |
- |
| 19 |
Hb_000016_060 |
0.1671925611 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 20 |
Hb_001217_010 |
0.1677762861 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |