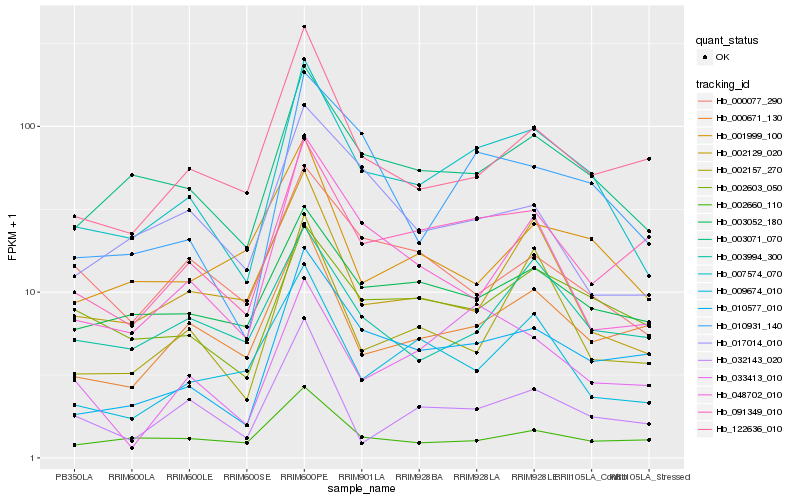
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007574_070 |
0.0 |
- |
- |
fatty acid desaturase, putative [Ricinus communis] |
| 2 |
Hb_003071_070 |
0.1188325755 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 3 |
Hb_010577_010 |
0.146243363 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 4 |
Hb_010931_140 |
0.1470223701 |
- |
- |
- |
| 5 |
Hb_091349_010 |
0.1591748125 |
- |
- |
hypothetical protein EUGRSUZ_C02287 [Eucalyptus grandis] |
| 6 |
Hb_002603_050 |
0.1612792397 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_003052_180 |
0.1621051393 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 8 |
Hb_000671_130 |
0.1681600248 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002157_270 |
0.1694532057 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 10 |
Hb_048702_010 |
0.1710808149 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_002129_020 |
0.173013195 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 12 |
Hb_122636_010 |
0.1743620978 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 13 |
Hb_009674_010 |
0.1771947287 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 14 |
Hb_003994_300 |
0.1778285799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002660_110 |
0.1812548895 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_032143_020 |
0.1832330075 |
- |
- |
hypothetical protein POPTR_0009s06310g [Populus trichocarpa] |
| 17 |
Hb_000077_290 |
0.1853266542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_033413_010 |
0.1859615413 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 19 |
Hb_001999_100 |
0.1861207816 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 20 |
Hb_017014_010 |
0.1861221262 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |