| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
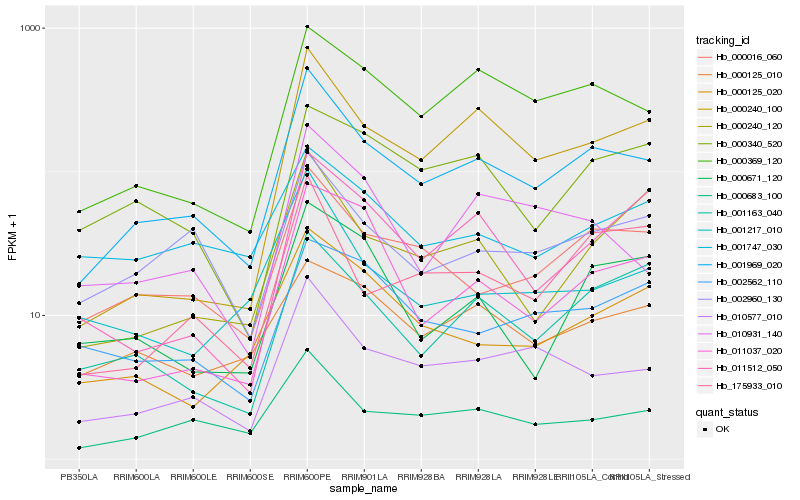
Hb_001969_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 2 |
Hb_000683_100 |
0.1180024178 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 3 |
Hb_010577_010 |
0.1189499758 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 4 |
Hb_000016_060 |
0.1198903482 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 5 |
Hb_000369_120 |
0.1332398815 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3-B [Jatropha curcas] |
| 6 |
Hb_002960_130 |
0.140871403 |
- |
- |
hypothetical protein B456_008G036100 [Gossypium raimondii] |
| 7 |
Hb_001163_040 |
0.1552364668 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 8 |
Hb_000125_020 |
0.1564949912 |
transcription factor |
TF Family: mTERF |
- |
| 9 |
Hb_175933_010 |
0.1570142874 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 10 |
Hb_010931_140 |
0.1600881522 |
- |
- |
- |
| 11 |
Hb_000240_100 |
0.1604472034 |
- |
- |
PREDICTED: ras-related protein Rab5-like [Eucalyptus grandis] |
| 12 |
Hb_001217_010 |
0.1612298914 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |
| 13 |
Hb_000125_010 |
0.1634007359 |
transcription factor |
TF Family: mTERF |
- |
| 14 |
Hb_000240_120 |
0.165016859 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 15 |
Hb_001747_030 |
0.1680402356 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 16 |
Hb_002562_110 |
0.1692084408 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000340_520 |
0.1716016579 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 18 |
Hb_000671_120 |
0.1716580034 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 19 |
Hb_011512_050 |
0.1733667745 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 20 |
Hb_011037_020 |
0.1754284504 |
- |
- |
unknown [Picea sitchensis] |