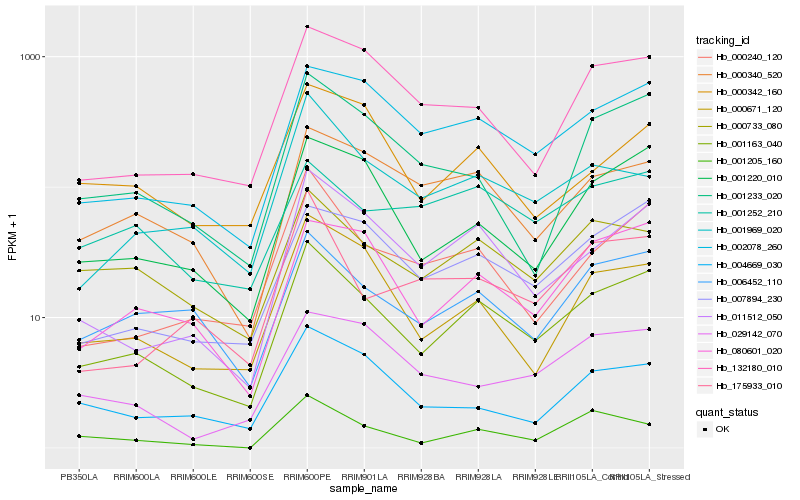
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001163_040 |
0.0 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 2 |
Hb_011512_050 |
0.1208528915 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 3 |
Hb_002078_260 |
0.1271547507 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_001220_010 |
0.1298889061 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 5 |
Hb_006452_110 |
0.1299223737 |
- |
- |
Ubiquinol-cytochrome c reductase complex 7.8 kDa protein, putative [Ricinus communis] |
| 6 |
Hb_007894_230 |
0.1307901735 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 7 |
Hb_080601_020 |
0.131102447 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 8 |
Hb_000733_080 |
0.1353218401 |
- |
- |
PREDICTED: uncharacterized protein LOC105646960 [Jatropha curcas] |
| 9 |
Hb_000671_120 |
0.1383491689 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 10 |
Hb_000240_120 |
0.1400125008 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 11 |
Hb_000340_520 |
0.1466745065 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 12 |
Hb_132180_010 |
0.1502082268 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 13 |
Hb_001233_020 |
0.1549523592 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 14 |
Hb_001969_020 |
0.1552364668 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 15 |
Hb_004669_030 |
0.1599581726 |
- |
- |
- |
| 16 |
Hb_001205_160 |
0.1600655275 |
- |
- |
hypothetical protein POPTR_0005s12560g [Populus trichocarpa] |
| 17 |
Hb_000342_160 |
0.1621266855 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 18 |
Hb_001252_210 |
0.1648266609 |
- |
- |
PREDICTED: glycine cleavage system H protein 2, mitochondrial [Jatropha curcas] |
| 19 |
Hb_175933_010 |
0.1679326043 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 20 |
Hb_029142_070 |
0.1684531039 |
- |
- |
- |