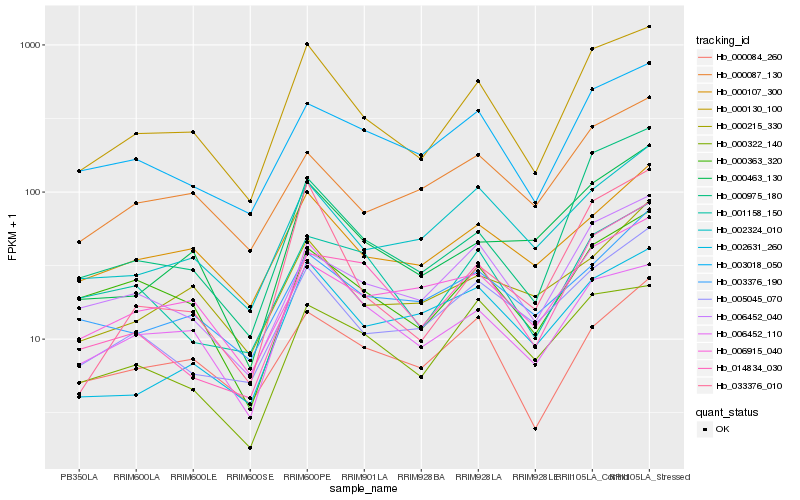
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_100 |
0.0 |
- |
- |
hypothetical protein CICLE_v10022662mg [Citrus clementina] |
| 2 |
Hb_003018_050 |
0.1117385641 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase Pin1 [Jatropha curcas] |
| 3 |
Hb_000975_180 |
0.1150602142 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 4 |
Hb_006452_040 |
0.1177159127 |
- |
- |
PREDICTED: signal recognition particle 9 kDa protein [Jatropha curcas] |
| 5 |
Hb_002324_010 |
0.1195791307 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 6 |
Hb_000107_300 |
0.1205425158 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-2 [Jatropha curcas] |
| 7 |
Hb_005045_070 |
0.1219760077 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 8 |
Hb_000363_320 |
0.123911192 |
- |
- |
PREDICTED: V-type proton ATPase subunit e1 [Jatropha curcas] |
| 9 |
Hb_006452_110 |
0.1246863218 |
- |
- |
Ubiquinol-cytochrome c reductase complex 7.8 kDa protein, putative [Ricinus communis] |
| 10 |
Hb_000084_260 |
0.127188807 |
- |
- |
PREDICTED: replication protein A 14 kDa subunit A-like [Jatropha curcas] |
| 11 |
Hb_001158_150 |
0.1285929222 |
- |
- |
hypothetical protein JCGZ_26407 [Jatropha curcas] |
| 12 |
Hb_000087_130 |
0.1291363336 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000463_130 |
0.1296193715 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_006915_040 |
0.1338661172 |
- |
- |
PREDICTED: prefoldin subunit 6 [Eucalyptus grandis] |
| 15 |
Hb_000322_140 |
0.1348578561 |
- |
- |
hypothetical protein RCOM_1047200 [Ricinus communis] |
| 16 |
Hb_000215_330 |
0.13565781 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 17 |
Hb_033376_010 |
0.1367647705 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 18 |
Hb_003376_190 |
0.1373925129 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 19 |
Hb_014834_030 |
0.1382525011 |
- |
- |
- |
| 20 |
Hb_002631_260 |
0.1384181875 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |