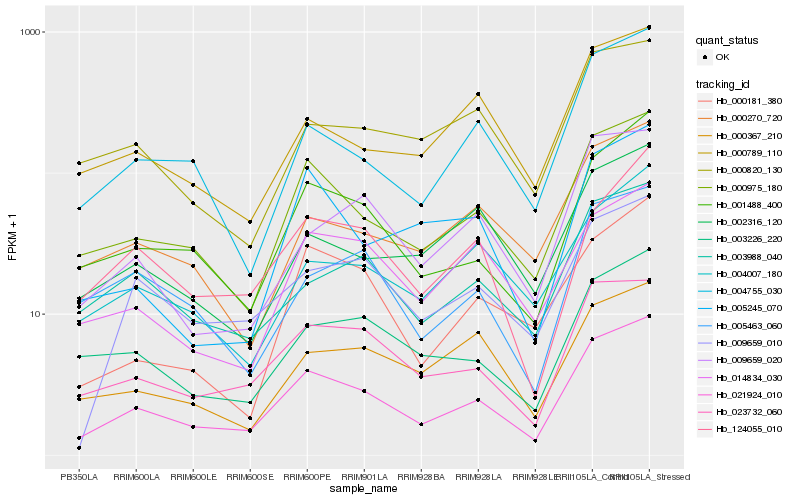
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021924_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 2 |
Hb_000975_180 |
0.0895838214 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 3 |
Hb_001488_400 |
0.1005433232 |
- |
- |
hypothetical protein POPTR_0018s12030g [Populus trichocarpa] |
| 4 |
Hb_023732_060 |
0.1121271716 |
- |
- |
PREDICTED: uncharacterized protein LOC105637567 [Jatropha curcas] |
| 5 |
Hb_004755_030 |
0.121689447 |
- |
- |
EG964 [Manihot glaziovii] |
| 6 |
Hb_003226_220 |
0.1278952066 |
- |
- |
hypothetical protein CICLE_v10022836mg [Citrus clementina] |
| 7 |
Hb_003988_040 |
0.1313017043 |
- |
- |
PREDICTED: uncharacterized protein LOC105633684 [Jatropha curcas] |
| 8 |
Hb_000270_720 |
0.1363550949 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 9 |
Hb_009659_010 |
0.137377276 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 9 [Vitis vinifera] |
| 10 |
Hb_000789_110 |
0.1379563457 |
- |
- |
RecName: Full=Profilin-1; AltName: Full=Pollen allergen Hev b 8.0101; AltName: Allergen=Hev b 8.0101 [Hevea brasiliensis] |
| 11 |
Hb_004007_180 |
0.1390712878 |
- |
- |
PREDICTED: uncharacterized protein LOC105639838 [Jatropha curcas] |
| 12 |
Hb_124055_010 |
0.1393386649 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 13 |
Hb_009659_020 |
0.1421177137 |
- |
- |
PREDICTED: uncharacterized protein LOC103870090 [Brassica rapa] |
| 14 |
Hb_005463_060 |
0.1427047753 |
- |
- |
PREDICTED: uncharacterized protein LOC105632515 [Jatropha curcas] |
| 15 |
Hb_014834_030 |
0.1450456227 |
- |
- |
- |
| 16 |
Hb_000367_210 |
0.1464139675 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 17 |
Hb_000181_380 |
0.1467210325 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 18 |
Hb_002316_120 |
0.1473586382 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 19 |
Hb_000820_130 |
0.1483800738 |
- |
- |
PREDICTED: uncharacterized protein LOC105769309 isoform X1 [Gossypium raimondii] |
| 20 |
Hb_005245_070 |
0.1486665926 |
- |
- |
PREDICTED: uncharacterized protein LOC105643658 [Jatropha curcas] |