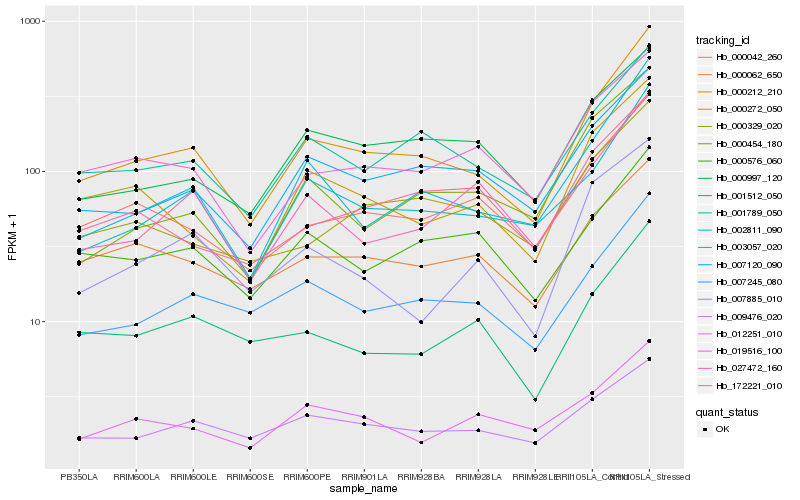
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_210 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_002811_090 |
0.0846486938 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 3 |
Hb_003057_020 |
0.0854208492 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |
| 4 |
Hb_001789_050 |
0.0914095894 |
- |
- |
60S ribosomal protein L36-2 [Morus notabilis] |
| 5 |
Hb_007120_090 |
0.1012051967 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 6 |
Hb_000042_260 |
0.1025508293 |
- |
- |
PREDICTED: uncharacterized protein LOC105632814 [Jatropha curcas] |
| 7 |
Hb_000272_050 |
0.1053319661 |
- |
- |
hypothetical protein JCGZ_03991 [Jatropha curcas] |
| 8 |
Hb_007245_080 |
0.1063601886 |
- |
- |
PREDICTED: uncharacterized protein LOC105644587 [Jatropha curcas] |
| 9 |
Hb_019516_100 |
0.1118780191 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 10 |
Hb_027472_160 |
0.1194534334 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 11 |
Hb_000997_120 |
0.1200993252 |
- |
- |
40S ribosomal protein S15E [Hevea brasiliensis] |
| 12 |
Hb_009476_020 |
0.1210977941 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 13 |
Hb_000329_020 |
0.1232469081 |
- |
- |
cytoplasmic ribosomal protein S13 [Panax ginseng] |
| 14 |
Hb_001512_050 |
0.1245821923 |
- |
- |
copine, putative [Ricinus communis] |
| 15 |
Hb_172221_010 |
0.1270409941 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |
| 16 |
Hb_000576_060 |
0.1273216415 |
- |
- |
truncated hemoglobin [Populus tremula x Populus tremuloides] |
| 17 |
Hb_000454_180 |
0.1273607005 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 18 |
Hb_012251_010 |
0.1282157515 |
- |
- |
- |
| 19 |
Hb_000062_650 |
0.1285627866 |
- |
- |
PREDICTED: uncharacterized protein LOC105628245 [Jatropha curcas] |
| 20 |
Hb_007885_010 |
0.1295002165 |
- |
- |
60S ribosomal protein L24-1 isoform 1 [Theobroma cacao] |