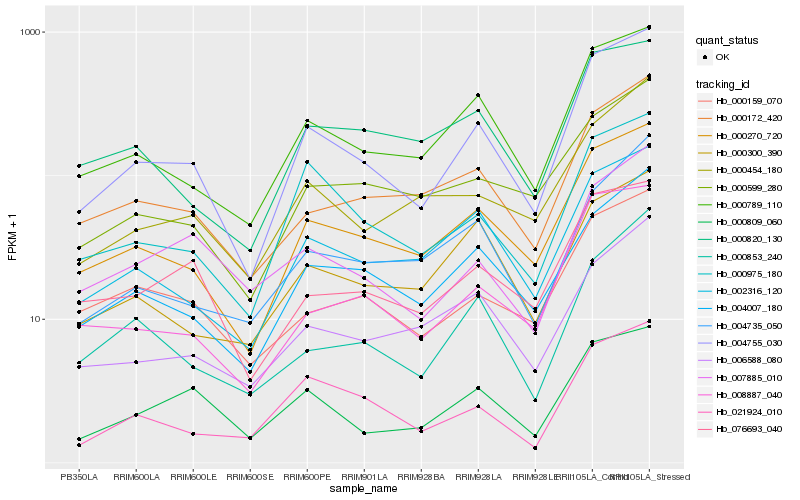
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004755_030 |
0.0 |
- |
- |
EG964 [Manihot glaziovii] |
| 2 |
Hb_000270_720 |
0.0826004775 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 3 |
Hb_000789_110 |
0.0944175773 |
- |
- |
RecName: Full=Profilin-1; AltName: Full=Pollen allergen Hev b 8.0101; AltName: Allergen=Hev b 8.0101 [Hevea brasiliensis] |
| 4 |
Hb_008887_040 |
0.1129831001 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_004007_180 |
0.1133820436 |
- |
- |
PREDICTED: uncharacterized protein LOC105639838 [Jatropha curcas] |
| 6 |
Hb_002316_120 |
0.1135876375 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 7 |
Hb_000975_180 |
0.1204896869 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 8 |
Hb_000172_420 |
0.1213159775 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 9 |
Hb_021924_010 |
0.121689447 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 10 |
Hb_000809_060 |
0.1218385969 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 11 |
Hb_000853_240 |
0.1275784406 |
- |
- |
- |
| 12 |
Hb_000454_180 |
0.129821066 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 13 |
Hb_000159_070 |
0.1314986673 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 14 |
Hb_076693_040 |
0.1315250355 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 15 |
Hb_000599_280 |
0.1326498948 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 16 |
Hb_006588_080 |
0.1346245629 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 17 |
Hb_004735_050 |
0.1349696463 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 18 |
Hb_007885_010 |
0.1395115085 |
- |
- |
60S ribosomal protein L24-1 isoform 1 [Theobroma cacao] |
| 19 |
Hb_000820_130 |
0.1403980568 |
- |
- |
PREDICTED: uncharacterized protein LOC105769309 isoform X1 [Gossypium raimondii] |
| 20 |
Hb_000300_390 |
0.140879698 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |