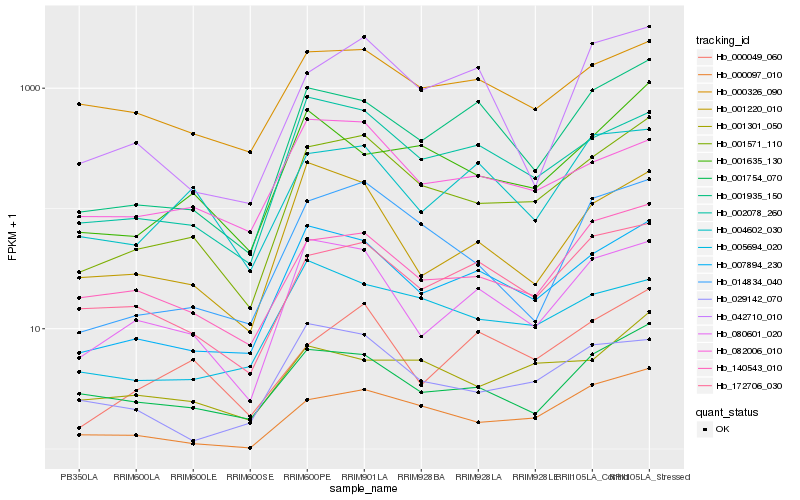
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001571_110 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L44-like [Gossypium raimondii] |
| 2 |
Hb_007894_230 |
0.1125022478 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 3 |
Hb_000097_010 |
0.1180272193 |
- |
- |
- |
| 4 |
Hb_014834_040 |
0.1252374374 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 5 |
Hb_140543_010 |
0.1366864194 |
- |
- |
- |
| 6 |
Hb_002078_260 |
0.1425666114 |
- |
- |
unknown [Populus trichocarpa] |
| 7 |
Hb_001935_150 |
0.1458988549 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 8 |
Hb_080601_020 |
0.1461468262 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 9 |
Hb_001754_070 |
0.1488099988 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 10 |
Hb_172706_030 |
0.1516539851 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |
| 11 |
Hb_001635_130 |
0.1551574926 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 12 |
Hb_001301_050 |
0.1586897074 |
- |
- |
- |
| 13 |
Hb_001220_010 |
0.1643757744 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 14 |
Hb_000326_090 |
0.1677840345 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein GRP1A-like [Nelumbo nucifera] |
| 15 |
Hb_000049_060 |
0.1684829457 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_005694_020 |
0.1701418264 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 17 |
Hb_029142_070 |
0.1713878499 |
- |
- |
- |
| 18 |
Hb_042710_010 |
0.1737592861 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 19 |
Hb_082006_010 |
0.174415305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004602_030 |
0.1754354324 |
- |
- |
histone H4 [Zea mays] |