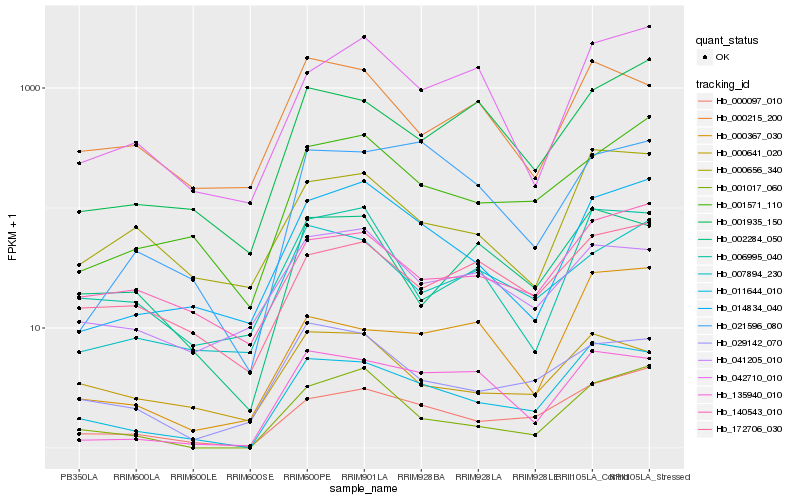
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011644_010 |
0.0 |
- |
- |
PREDICTED: probable choline-phosphate cytidylyltransferase [Elaeis guineensis] |
| 2 |
Hb_000097_010 |
0.1243530062 |
- |
- |
- |
| 3 |
Hb_135940_010 |
0.1580673008 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 4 |
Hb_001017_060 |
0.1662768667 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 5 |
Hb_029142_070 |
0.1669128828 |
- |
- |
- |
| 6 |
Hb_000656_340 |
0.1674065028 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 7 |
Hb_014834_040 |
0.1677826064 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 8 |
Hb_002284_050 |
0.1712571081 |
- |
- |
hypothetical protein POPTR_0002s00740g [Populus trichocarpa] |
| 9 |
Hb_001571_110 |
0.1788531168 |
- |
- |
PREDICTED: 60S ribosomal protein L44-like [Gossypium raimondii] |
| 10 |
Hb_140543_010 |
0.1790446739 |
- |
- |
- |
| 11 |
Hb_006995_040 |
0.1799329288 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 12 |
Hb_172706_030 |
0.181059378 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |
| 13 |
Hb_001935_150 |
0.1838872694 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 14 |
Hb_000641_020 |
0.185636109 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 15 |
Hb_000367_030 |
0.1864124432 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |
| 16 |
Hb_021596_080 |
0.1866047583 |
- |
- |
50S ribosomal protein L33A [Hevea brasiliensis] |
| 17 |
Hb_042710_010 |
0.1883340326 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 18 |
Hb_000215_200 |
0.1887629564 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 19 |
Hb_007894_230 |
0.1903962465 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 20 |
Hb_041205_010 |
0.191064495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |