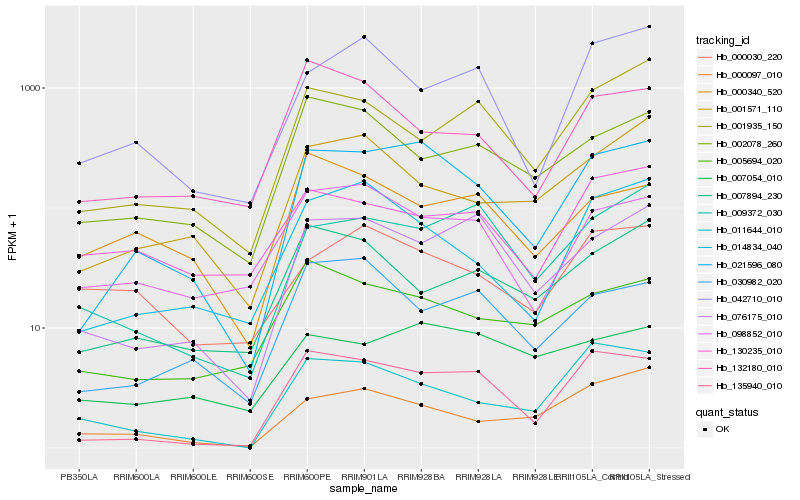
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021596_080 |
0.0 |
- |
- |
50S ribosomal protein L33A [Hevea brasiliensis] |
| 2 |
Hb_135940_010 |
0.1416226079 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 3 |
Hb_014834_040 |
0.1661266789 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 4 |
Hb_076175_010 |
0.1758005394 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB7 [Cucumis melo] |
| 5 |
Hb_098852_010 |
0.1789085789 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 6 |
Hb_011644_010 |
0.1866047583 |
- |
- |
PREDICTED: probable choline-phosphate cytidylyltransferase [Elaeis guineensis] |
| 7 |
Hb_000097_010 |
0.1866249834 |
- |
- |
- |
| 8 |
Hb_005694_020 |
0.1881642337 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 9 |
Hb_009372_030 |
0.189917884 |
- |
- |
50S ribosomal protein L6, putative [Ricinus communis] |
| 10 |
Hb_007054_010 |
0.1925774759 |
- |
- |
PREDICTED: WD repeat-containing protein 82 [Jatropha curcas] |
| 11 |
Hb_001571_110 |
0.1937365299 |
- |
- |
PREDICTED: 60S ribosomal protein L44-like [Gossypium raimondii] |
| 12 |
Hb_030982_020 |
0.1953183369 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_042710_010 |
0.1974048691 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 14 |
Hb_002078_260 |
0.1974532751 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_001935_150 |
0.2017197687 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 16 |
Hb_007894_230 |
0.2081400112 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 17 |
Hb_132180_010 |
0.208854188 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 18 |
Hb_000030_220 |
0.2090067908 |
- |
- |
PREDICTED: uncharacterized protein LOC105650405 [Jatropha curcas] |
| 19 |
Hb_130235_010 |
0.2139815774 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 20 |
Hb_000340_520 |
0.2147592585 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |