Hb_002740_010
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig2740: 78471-79546 |
| Sequence |   |
Annotation
kegg
| ID | gmx:100792684 |
|---|---|
| description | hydrophobic protein LTI6A-like |
nr
| ID | ADC45381.1 |
|---|---|
| description | stress-induced hydrophobic peptide [Cleistogenes songorica] |
swissprot
| ID | Q8H5T6 |
|---|---|
| description | Hydrophobic protein LTI6A OS=Oryza sativa subsp. japonica GN=LTI6A PE=2 SV=1 |
trembl
| ID | D6C5C2 |
|---|---|
| description | Stress-induced hydrophobic peptide OS=Cleistogenes songorica PE=2 SV=1 |
Gene Ontology
| ID | GO:0016021 |
|---|---|
| description | stress-induced hydrophobic peptide |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_28659: 78339-79781 , PASA_asmbl_28660: 78764-79781 |
|---|---|
| cDNA (Sanger) (ID:Location) |
002_B13.ab1: 78449-79650 , 002_M12.ab1: 78505-79650 , 004_J02.ab1: 78505-79651 , 005_J10.ab1: 78443-79650 , 008_E07.ab1: 78451-79650 , 010_I18.ab1: 78445-79652 , 015_C20.ab1: 78585-79650 , 022_C04.ab1: 78459-79645 , 024_F07.ab1: 78529-79650 , 024_L04.ab1: 78457-79655 , 027_P04.ab1: 78443-79653 , 032_L01.ab1: 78442-79650 , 038_B02.ab1: 78445-79781 , 039_H08.ab1: 78450-79650 , 039_J19.ab1: 78585-79650 , 043_H08.ab1: 78453-79472 , 046_K04.ab1: 78505-79650 , 050_L06.ab1: 78455-79650 , 050_O05.ab1: 78445-79652 |
Similar expressed genes (Top20)
Gene co-expression network
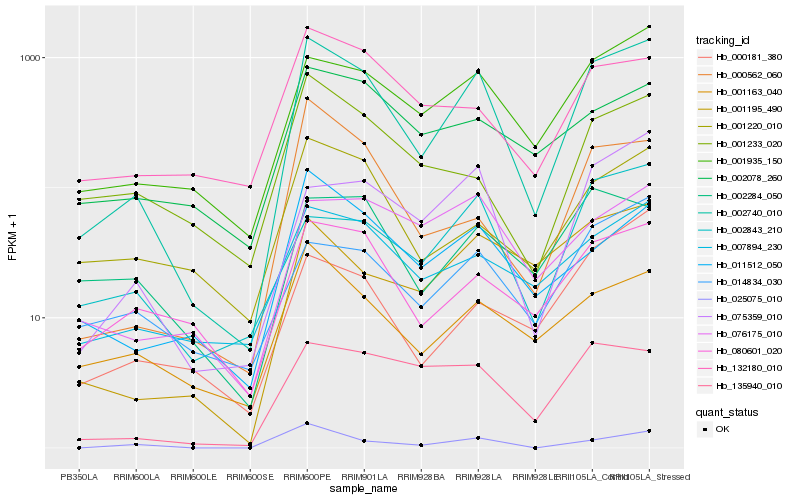
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 86.8258 | 4.67271 | 11.4863 | 1427.68 | 40.147 | 779.143 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 924.879 | 1377.15 | 796.587 | 170.371 | 60.0907 |

