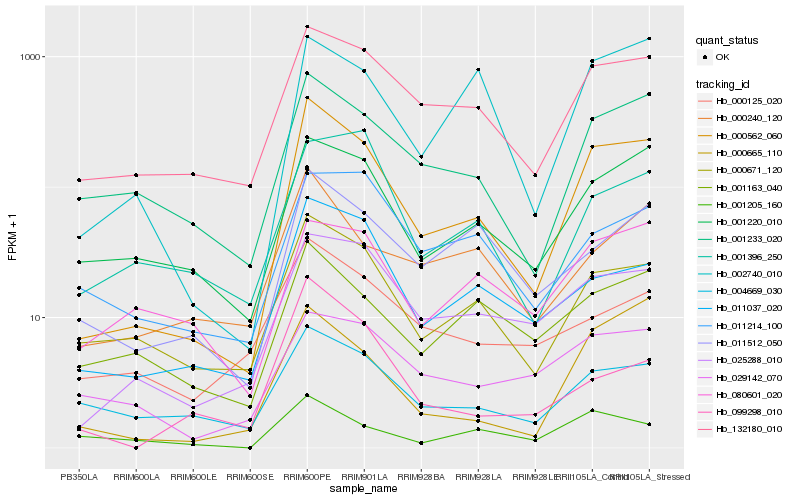
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000562_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105169741 [Sesamum indicum] |
| 2 |
Hb_132180_010 |
0.1705545481 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 3 |
Hb_000671_120 |
0.1739050494 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 4 |
Hb_025288_010 |
0.1756989876 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 5 |
Hb_001233_020 |
0.1784445555 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 6 |
Hb_011037_020 |
0.1829612322 |
- |
- |
unknown [Picea sitchensis] |
| 7 |
Hb_001220_010 |
0.1850761981 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 8 |
Hb_002740_010 |
0.187082913 |
- |
- |
stress-induced hydrophobic peptide [Cleistogenes songorica] |
| 9 |
Hb_000665_110 |
0.1915775345 |
- |
- |
Ribosomal protein S11-beta [Theobroma cacao] |
| 10 |
Hb_011512_050 |
0.198399219 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 11 |
Hb_000240_120 |
0.2076218412 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 12 |
Hb_004669_030 |
0.2088650995 |
- |
- |
- |
| 13 |
Hb_001163_040 |
0.2130669958 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 14 |
Hb_001396_250 |
0.2219225248 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 15 |
Hb_099298_010 |
0.2229387681 |
- |
- |
- |
| 16 |
Hb_001205_160 |
0.2230025995 |
- |
- |
hypothetical protein POPTR_0005s12560g [Populus trichocarpa] |
| 17 |
Hb_029142_070 |
0.2233753407 |
- |
- |
- |
| 18 |
Hb_011214_100 |
0.2233785787 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 19 |
Hb_080601_020 |
0.2235933488 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 20 |
Hb_000125_020 |
0.2236998423 |
transcription factor |
TF Family: mTERF |
- |