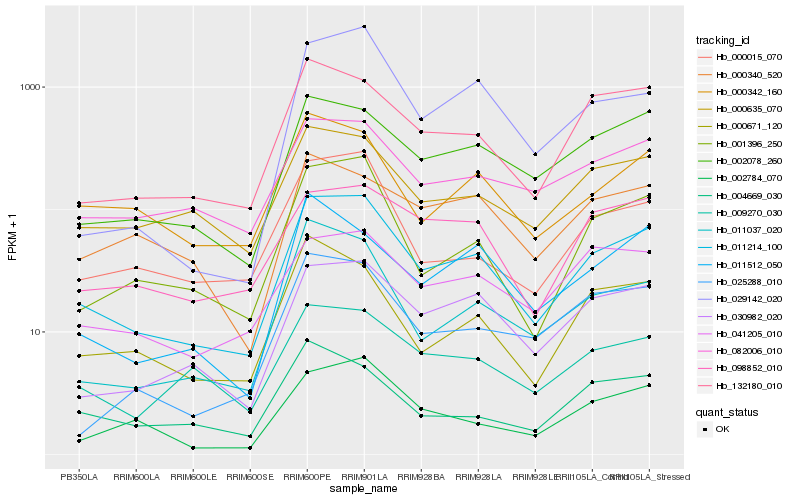
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011214_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 2 |
Hb_001396_250 |
0.1131961311 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 3 |
Hb_030982_020 |
0.1142670177 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_132180_010 |
0.1164062493 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 5 |
Hb_002078_260 |
0.1178081395 |
- |
- |
unknown [Populus trichocarpa] |
| 6 |
Hb_000015_070 |
0.1277502469 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 7 |
Hb_000342_160 |
0.1307483427 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 8 |
Hb_000671_120 |
0.131342109 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 9 |
Hb_011037_020 |
0.1314012435 |
- |
- |
unknown [Picea sitchensis] |
| 10 |
Hb_002784_070 |
0.132900929 |
- |
- |
BnaUnng01470D [Brassica napus] |
| 11 |
Hb_000635_070 |
0.1329733755 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 12 |
Hb_082006_010 |
0.1351473738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009270_030 |
0.1363347227 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 14 |
Hb_098852_010 |
0.1407285428 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 15 |
Hb_041205_010 |
0.143248686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_025288_010 |
0.147016924 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 17 |
Hb_011512_050 |
0.1477036447 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 18 |
Hb_029142_020 |
0.1500760166 |
- |
- |
hypothetical protein POPTR_0004s23900g [Populus trichocarpa] |
| 19 |
Hb_004669_030 |
0.1503392794 |
- |
- |
- |
| 20 |
Hb_000340_520 |
0.1526958169 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |