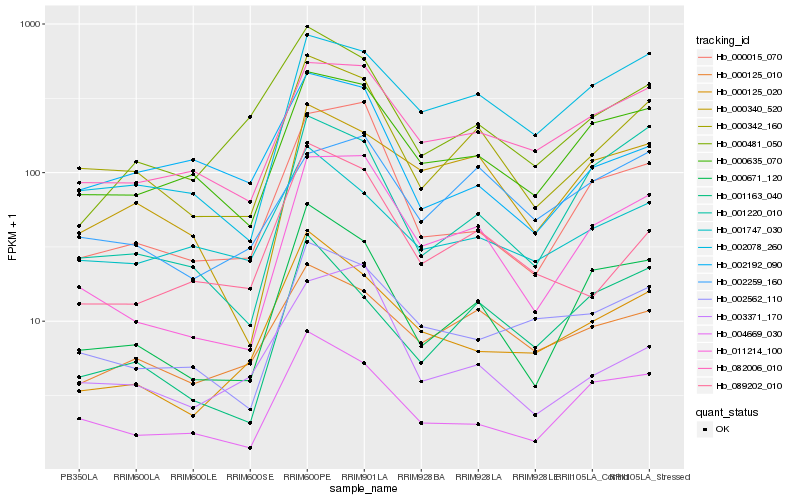
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000342_160 |
0.0 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 2 |
Hb_000671_120 |
0.1191088858 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 3 |
Hb_000635_070 |
0.1241927643 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 4 |
Hb_011214_100 |
0.1307483427 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 5 |
Hb_082006_010 |
0.1314138274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000125_010 |
0.1340673124 |
transcription factor |
TF Family: mTERF |
- |
| 7 |
Hb_004669_030 |
0.1346767583 |
- |
- |
- |
| 8 |
Hb_001747_030 |
0.1410552936 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 9 |
Hb_089202_010 |
0.1448290329 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 10 |
Hb_002562_110 |
0.148230225 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000340_520 |
0.1485506197 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 12 |
Hb_000481_050 |
0.1498781713 |
- |
- |
PREDICTED: uncharacterized protein LOC105636546 [Jatropha curcas] |
| 13 |
Hb_000015_070 |
0.1537956827 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 14 |
Hb_001220_010 |
0.1550006756 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 15 |
Hb_002192_090 |
0.1560988666 |
- |
- |
hypothetical protein AMTR_s00029p00055000 [Amborella trichopoda] |
| 16 |
Hb_002078_260 |
0.1579492447 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_000125_020 |
0.1584085488 |
transcription factor |
TF Family: mTERF |
- |
| 18 |
Hb_001163_040 |
0.1621266855 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 19 |
Hb_002259_160 |
0.163554932 |
- |
- |
hypothetical protein PSEUBRA_SCAF14g01508 [Pseudozyma brasiliensis GHG001] |
| 20 |
Hb_003371_170 |
0.1650583857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |