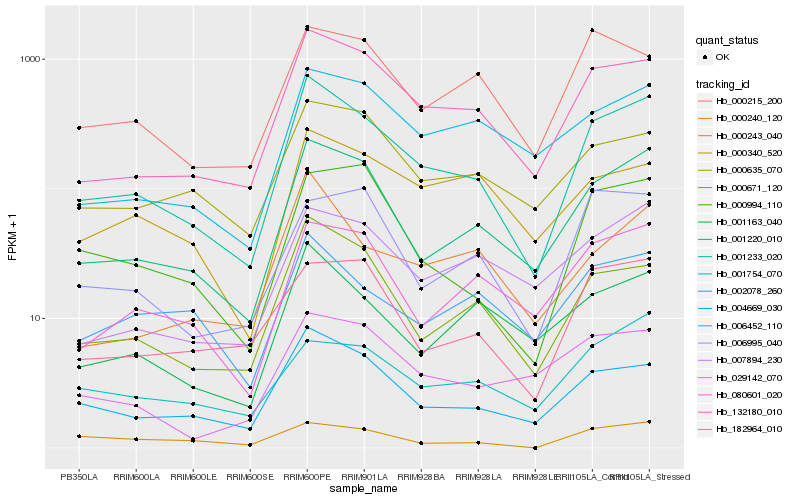
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001233_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 2 |
Hb_001220_010 |
0.1018094824 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 3 |
Hb_132180_010 |
0.1060952544 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 4 |
Hb_004669_030 |
0.1132288881 |
- |
- |
- |
| 5 |
Hb_000671_120 |
0.120180216 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 6 |
Hb_080601_020 |
0.1520489428 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 7 |
Hb_000994_110 |
0.1527583345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001163_040 |
0.1549523592 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 9 |
Hb_000635_070 |
0.1589025383 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 10 |
Hb_182964_010 |
0.1595078637 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |
| 11 |
Hb_006452_110 |
0.1608725684 |
- |
- |
Ubiquinol-cytochrome c reductase complex 7.8 kDa protein, putative [Ricinus communis] |
| 12 |
Hb_000240_120 |
0.1614140203 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 13 |
Hb_002078_260 |
0.1627448288 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_007894_230 |
0.1643792209 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 15 |
Hb_000215_200 |
0.1658360025 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 16 |
Hb_001754_070 |
0.1698626139 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 17 |
Hb_029142_070 |
0.1710014707 |
- |
- |
- |
| 18 |
Hb_000340_520 |
0.171166781 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 19 |
Hb_006995_040 |
0.1719866104 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 20 |
Hb_000243_040 |
0.175617148 |
- |
- |
- |