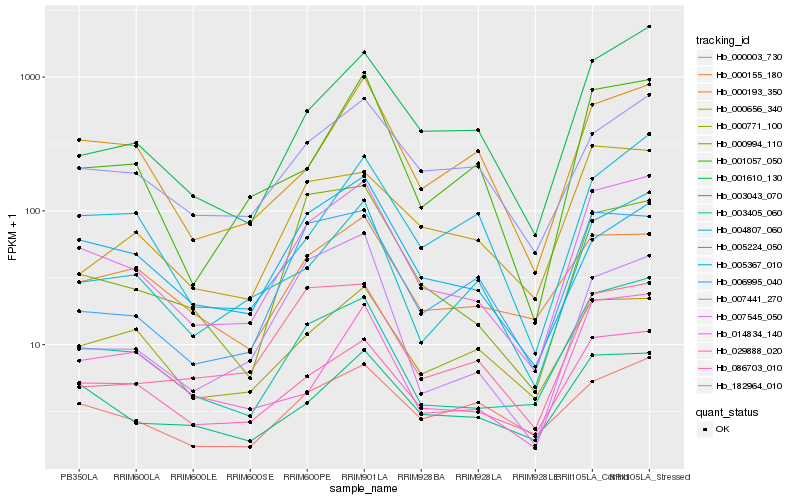
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014834_140 |
0.0 |
- |
- |
Uncharacterized protein TCM_015942 [Theobroma cacao] |
| 2 |
Hb_003405_060 |
0.0891232563 |
- |
- |
- |
| 3 |
Hb_086703_010 |
0.1065109572 |
- |
- |
PREDICTED: F-box protein At5g52880 [Jatropha curcas] |
| 4 |
Hb_004807_060 |
0.1215879645 |
- |
- |
- |
| 5 |
Hb_001610_130 |
0.1276214566 |
- |
- |
40S ribosomal protein S30 [Medicago truncatula] |
| 6 |
Hb_003043_070 |
0.134207308 |
- |
- |
- |
| 7 |
Hb_001057_050 |
0.1352162517 |
- |
- |
hypothetical protein POPTR_0003s15960g, partial [Populus trichocarpa] |
| 8 |
Hb_007441_270 |
0.137829869 |
- |
- |
PREDICTED: 40S ribosomal protein S11 [Jatropha curcas] |
| 9 |
Hb_006995_040 |
0.1400638498 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 10 |
Hb_000193_350 |
0.1413899756 |
- |
- |
eukaryotic translation initiation factor 5A isoform VI [Hevea brasiliensis] |
| 11 |
Hb_000994_110 |
0.1451795963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000656_340 |
0.1491476271 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 13 |
Hb_029888_020 |
0.149589884 |
- |
- |
hypothetical protein PHAVU_007G268100g [Phaseolus vulgaris] |
| 14 |
Hb_000771_100 |
0.1539845149 |
- |
- |
Os06g0527300 [Oryza sativa Japonica Group] |
| 15 |
Hb_000155_180 |
0.1567657293 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 16 |
Hb_005224_050 |
0.1582216804 |
- |
- |
PREDICTED: uncharacterized protein LOC105632350 [Jatropha curcas] |
| 17 |
Hb_007545_050 |
0.1586717358 |
- |
- |
- |
| 18 |
Hb_000003_730 |
0.1591250693 |
- |
- |
DNA repair/transcription protein met18/mms19, putative [Ricinus communis] |
| 19 |
Hb_182964_010 |
0.159393397 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005367_010 |
0.1624588594 |
- |
- |
- |