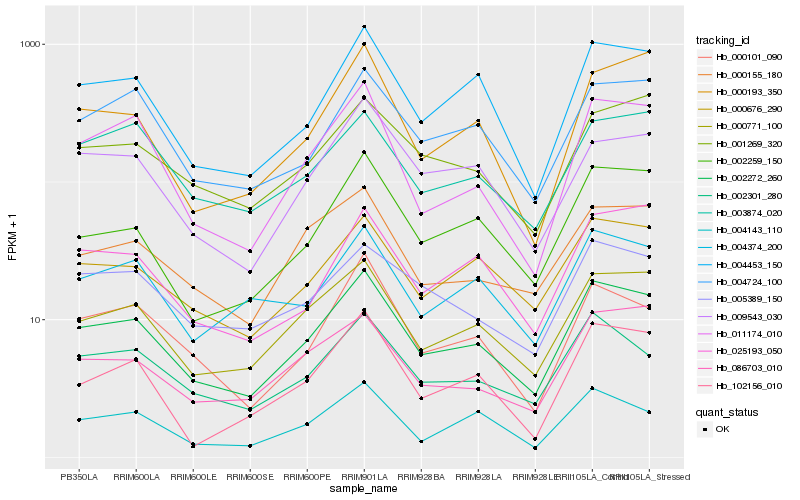
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002272_260 |
0.0 |
- |
- |
- |
| 2 |
Hb_000771_100 |
0.068116764 |
- |
- |
Os06g0527300 [Oryza sativa Japonica Group] |
| 3 |
Hb_004453_150 |
0.0819029783 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 4 |
Hb_000676_290 |
0.0868957693 |
- |
- |
PREDICTED: glutaredoxin-C3 [Jatropha curcas] |
| 5 |
Hb_002301_280 |
0.0906647638 |
transcription factor |
TF Family: SBP |
hypothetical protein JCGZ_21462 [Jatropha curcas] |
| 6 |
Hb_002259_150 |
0.0913126468 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 7 |
Hb_011174_010 |
0.0933882538 |
- |
- |
hypothetical protein JCGZ_10122 [Jatropha curcas] |
| 8 |
Hb_004724_100 |
0.0977761729 |
- |
- |
60S ribosomal protein L10aA [Hevea brasiliensis] |
| 9 |
Hb_000101_090 |
0.0984569678 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Camelina sativa] |
| 10 |
Hb_000155_180 |
0.0994027704 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 11 |
Hb_004143_110 |
0.1017267759 |
- |
- |
PREDICTED: uncharacterized protein LOC105630719 isoform X2 [Jatropha curcas] |
| 12 |
Hb_009543_030 |
0.1021560157 |
- |
- |
prefoldin subunit 1 [Jatropha curcas] |
| 13 |
Hb_025193_050 |
0.1034166129 |
- |
- |
PREDICTED: uncharacterized protein LOC105647290 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001269_320 |
0.1041616611 |
- |
- |
40S ribosomal protein S12A [Hevea brasiliensis] |
| 15 |
Hb_004374_200 |
0.1052989876 |
- |
- |
putative GMP synthase [Oxybasis rubra] |
| 16 |
Hb_086703_010 |
0.1068834396 |
- |
- |
PREDICTED: F-box protein At5g52880 [Jatropha curcas] |
| 17 |
Hb_102156_010 |
0.1078198154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000193_350 |
0.1102963428 |
- |
- |
eukaryotic translation initiation factor 5A isoform VI [Hevea brasiliensis] |
| 19 |
Hb_005389_150 |
0.1104024015 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003874_020 |
0.111123876 |
- |
- |
hypothetical protein POPTR_0012s02280g [Populus trichocarpa] |