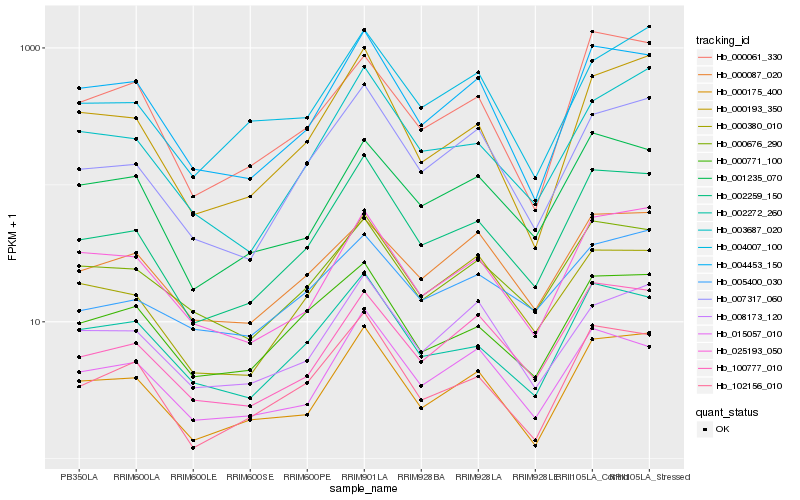
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_150 |
0.0 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 2 |
Hb_007317_060 |
0.0801520163 |
- |
- |
60S ribosomal protein L10aB [Hevea brasiliensis] |
| 3 |
Hb_002272_260 |
0.0913126468 |
- |
- |
- |
| 4 |
Hb_102156_010 |
0.0917832387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_100777_010 |
0.0937428459 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 19a-like [Populus euphratica] |
| 6 |
Hb_004453_150 |
0.0946458814 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 7 |
Hb_000193_350 |
0.0986389191 |
- |
- |
eukaryotic translation initiation factor 5A isoform VI [Hevea brasiliensis] |
| 8 |
Hb_000175_400 |
0.0995321461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003687_020 |
0.1006975462 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 10 |
Hb_000771_100 |
0.1030909464 |
- |
- |
Os06g0527300 [Oryza sativa Japonica Group] |
| 11 |
Hb_025193_050 |
0.1040132552 |
- |
- |
PREDICTED: uncharacterized protein LOC105647290 isoform X1 [Jatropha curcas] |
| 12 |
Hb_015057_010 |
0.1046197222 |
- |
- |
PREDICTED: uncharacterized exonuclease domain-containing protein At3g15140 [Jatropha curcas] |
| 13 |
Hb_000676_290 |
0.1049634461 |
- |
- |
PREDICTED: glutaredoxin-C3 [Jatropha curcas] |
| 14 |
Hb_001235_070 |
0.1051274111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000380_010 |
0.1072167831 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X3 [Jatropha curcas] |
| 16 |
Hb_004007_100 |
0.1181886323 |
- |
- |
PREDICTED: 60S ribosomal protein L26-1 [Jatropha curcas] |
| 17 |
Hb_005400_030 |
0.118220389 |
- |
- |
PREDICTED: LYR motif-containing protein At3g19508 [Jatropha curcas] |
| 18 |
Hb_008173_120 |
0.1188192038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000061_330 |
0.1203161951 |
- |
- |
eukaryotic translation initiation factor 5A isoform VII [Hevea brasiliensis] |
| 20 |
Hb_000087_020 |
0.1207857134 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase 2, mitochondrial [Jatropha curcas] |