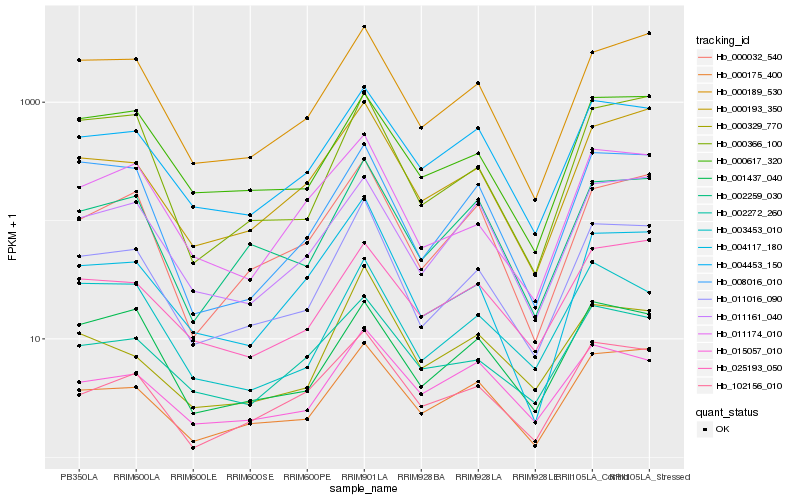
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011016_090 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L24, chloroplastic [Populus euphratica] |
| 2 |
Hb_000189_530 |
0.088741874 |
- |
- |
40S ribosomal protein S24B [Hevea brasiliensis] |
| 3 |
Hb_000175_400 |
0.0924639966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000193_350 |
0.0948611183 |
- |
- |
eukaryotic translation initiation factor 5A isoform VI [Hevea brasiliensis] |
| 5 |
Hb_002259_030 |
0.0951110235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004453_150 |
0.0968188154 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 7 |
Hb_000032_540 |
0.0983702274 |
- |
- |
PREDICTED: mediator-associated protein 3-like [Jatropha curcas] |
| 8 |
Hb_004117_180 |
0.1038262802 |
- |
- |
- |
| 9 |
Hb_011174_010 |
0.1100276275 |
- |
- |
hypothetical protein JCGZ_10122 [Jatropha curcas] |
| 10 |
Hb_102156_010 |
0.110566416 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000366_100 |
0.111094133 |
- |
- |
60S ribosomal protein L7aA [Hevea brasiliensis] |
| 12 |
Hb_000617_320 |
0.1130195093 |
- |
- |
40S ribosomal protein S21B [Hevea brasiliensis] |
| 13 |
Hb_011161_040 |
0.115189448 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002272_260 |
0.1165683256 |
- |
- |
- |
| 15 |
Hb_025193_050 |
0.1169368116 |
- |
- |
PREDICTED: uncharacterized protein LOC105647290 isoform X1 [Jatropha curcas] |
| 16 |
Hb_008016_010 |
0.1180276187 |
- |
- |
PREDICTED: protein LURP-one-related 15-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001437_040 |
0.1202914644 |
- |
- |
PREDICTED: ATPase GET3-like [Vitis vinifera] |
| 18 |
Hb_003453_010 |
0.1218154606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000329_770 |
0.121958116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_015057_010 |
0.1239087057 |
- |
- |
PREDICTED: uncharacterized exonuclease domain-containing protein At3g15140 [Jatropha curcas] |