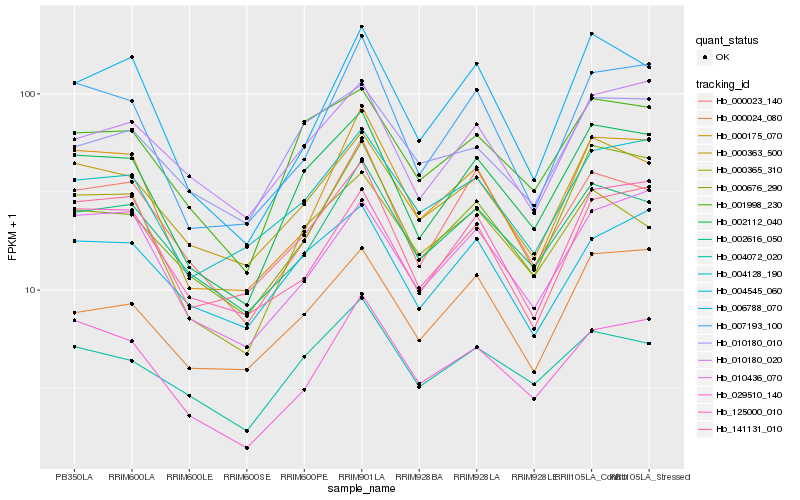
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002112_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 2 |
Hb_001998_230 |
0.0601944996 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 3 |
Hb_000363_500 |
0.0818123133 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004072_020 |
0.0850775704 |
- |
- |
PREDICTED: uncharacterized protein LOC105635274 [Jatropha curcas] |
| 5 |
Hb_000676_290 |
0.0858799228 |
- |
- |
PREDICTED: glutaredoxin-C3 [Jatropha curcas] |
| 6 |
Hb_002616_050 |
0.0874351518 |
- |
- |
PREDICTED: traB domain-containing protein [Jatropha curcas] |
| 7 |
Hb_004128_190 |
0.0898931217 |
- |
- |
PREDICTED: dirigent protein 17-like [Jatropha curcas] |
| 8 |
Hb_007193_100 |
0.0910479569 |
- |
- |
syntaxin family protein [Populus trichocarpa] |
| 9 |
Hb_000023_140 |
0.0913519462 |
- |
- |
PREDICTED: cadmium-induced protein AS8 isoform X1 [Jatropha curcas] |
| 10 |
Hb_010180_020 |
0.0914990888 |
- |
- |
hypothetical protein glysoja_030394 [Glycine soja] |
| 11 |
Hb_000365_310 |
0.0919406723 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 12 |
Hb_141131_010 |
0.0922689702 |
- |
- |
Diphthamide biosynthesis protein, putative [Ricinus communis] |
| 13 |
Hb_010436_070 |
0.0923585171 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 4-like [Nicotiana tomentosiformis] |
| 14 |
Hb_029510_140 |
0.0931542938 |
- |
- |
PREDICTED: uncharacterized protein LOC105635017 [Jatropha curcas] |
| 15 |
Hb_000024_080 |
0.093265606 |
- |
- |
PREDICTED: probable complex I intermediate-associated protein 30 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004545_060 |
0.0932870309 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 6B [Jatropha curcas] |
| 17 |
Hb_000175_070 |
0.0942366543 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 11 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006788_070 |
0.0975021615 |
- |
- |
2-methyl-6-geranylgeranylbenzoquinone methyltranferase [Hevea brasiliensis] |
| 19 |
Hb_010180_010 |
0.0985714597 |
- |
- |
PREDICTED: uncharacterized protein LOC105631104 [Jatropha curcas] |
| 20 |
Hb_125000_010 |
0.1001513485 |
- |
- |
PREDICTED: protein SCO1 homolog 2, mitochondrial [Jatropha curcas] |