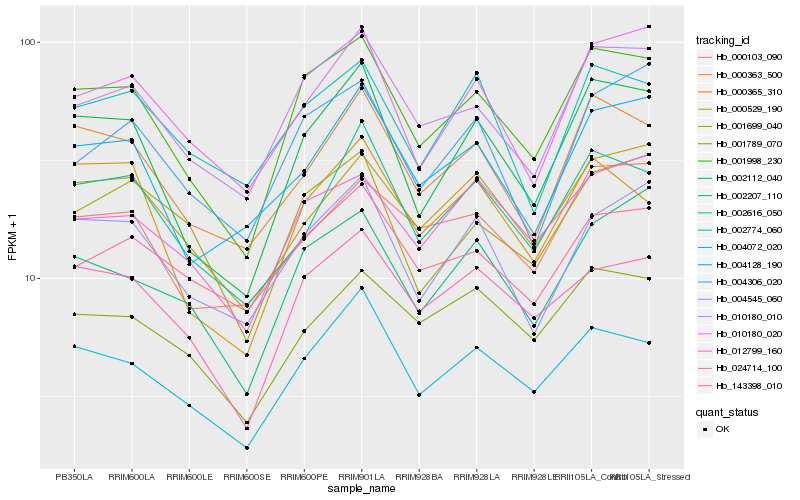
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001998_230 |
0.0 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 2 |
Hb_010180_010 |
0.0599137336 |
- |
- |
PREDICTED: uncharacterized protein LOC105631104 [Jatropha curcas] |
| 3 |
Hb_002112_040 |
0.0601944996 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 4 |
Hb_004072_020 |
0.0702684722 |
- |
- |
PREDICTED: uncharacterized protein LOC105635274 [Jatropha curcas] |
| 5 |
Hb_024714_100 |
0.0708433765 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000363_500 |
0.0825809677 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_012799_160 |
0.0846010987 |
- |
- |
PREDICTED: ADP-ribosylation factor-related protein 1-like [Jatropha curcas] |
| 8 |
Hb_002207_110 |
0.0870164471 |
- |
- |
- |
| 9 |
Hb_000529_190 |
0.0887049169 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1-like [Jatropha curcas] |
| 10 |
Hb_004545_060 |
0.0890910932 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 6B [Jatropha curcas] |
| 11 |
Hb_010180_020 |
0.0905127399 |
- |
- |
hypothetical protein glysoja_030394 [Glycine soja] |
| 12 |
Hb_001699_040 |
0.0907972614 |
- |
- |
PREDICTED: uncharacterized protein LOC105636529 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001789_070 |
0.0916754793 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 12 [Jatropha curcas] |
| 14 |
Hb_002616_050 |
0.0926429144 |
- |
- |
PREDICTED: traB domain-containing protein [Jatropha curcas] |
| 15 |
Hb_004128_190 |
0.0934097939 |
- |
- |
PREDICTED: dirigent protein 17-like [Jatropha curcas] |
| 16 |
Hb_000365_310 |
0.0946283983 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 17 |
Hb_004306_020 |
0.0952537882 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 6 [Jatropha curcas] |
| 18 |
Hb_000103_090 |
0.0959225605 |
- |
- |
PREDICTED: protein RDM1 [Jatropha curcas] |
| 19 |
Hb_143398_010 |
0.0965364247 |
- |
- |
PREDICTED: peter Pan-like protein [Jatropha curcas] |
| 20 |
Hb_002774_060 |
0.098395103 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X2 [Jatropha curcas] |