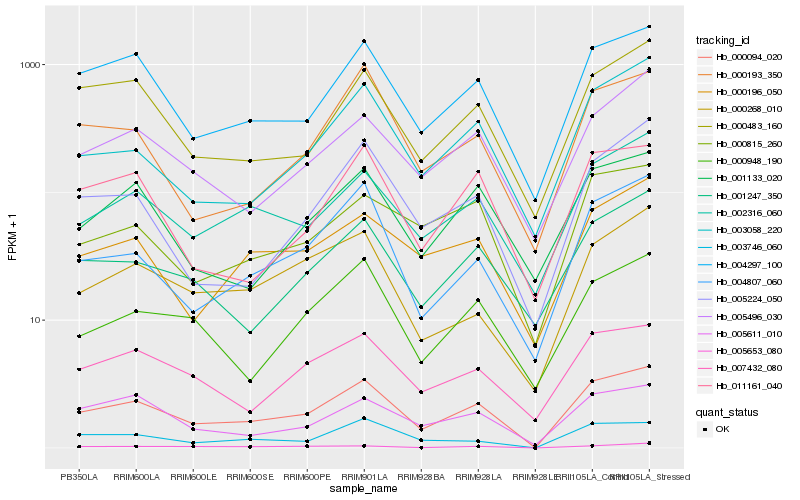
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_020 |
0.0 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 2 |
Hb_004297_100 |
0.110319432 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 3 |
Hb_002316_060 |
0.1170053538 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 4 |
Hb_004807_060 |
0.1245351566 |
- |
- |
- |
| 5 |
Hb_003746_060 |
0.1297738073 |
- |
- |
PREDICTED: DTW domain-containing protein 2 [Jatropha curcas] |
| 6 |
Hb_005611_010 |
0.1311852344 |
- |
- |
PREDICTED: protein prune homolog [Jatropha curcas] |
| 7 |
Hb_005496_030 |
0.1315405392 |
- |
- |
PREDICTED: 40S ribosomal protein S29-like [Malus domestica] |
| 8 |
Hb_000483_160 |
0.1322279657 |
- |
- |
PREDICTED: 60S ribosomal protein L34 [Jatropha curcas] |
| 9 |
Hb_000196_050 |
0.1326008941 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 10 |
Hb_003058_220 |
0.1332733693 |
- |
- |
Pre-mRNA branch site protein p14, putative [Ricinus communis] |
| 11 |
Hb_001247_350 |
0.1333968268 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 11 [Jatropha curcas] |
| 12 |
Hb_007432_080 |
0.1343575568 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1-like [Jatropha curcas] |
| 13 |
Hb_000815_260 |
0.13460741 |
- |
- |
50S ribosomal protein L32pA [Hevea brasiliensis] |
| 14 |
Hb_005653_080 |
0.1375646177 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000948_190 |
0.1380943082 |
transcription factor |
TF Family: GNAT |
Glucosamine 6-phosphate N-acetyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000268_010 |
0.1393349879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001133_020 |
0.1398300708 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase complex SCF subunit sconB [Jatropha curcas] |
| 18 |
Hb_005224_050 |
0.1411146113 |
- |
- |
PREDICTED: uncharacterized protein LOC105632350 [Jatropha curcas] |
| 19 |
Hb_000193_350 |
0.1420258904 |
- |
- |
eukaryotic translation initiation factor 5A isoform VI [Hevea brasiliensis] |
| 20 |
Hb_011161_040 |
0.1422426455 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |