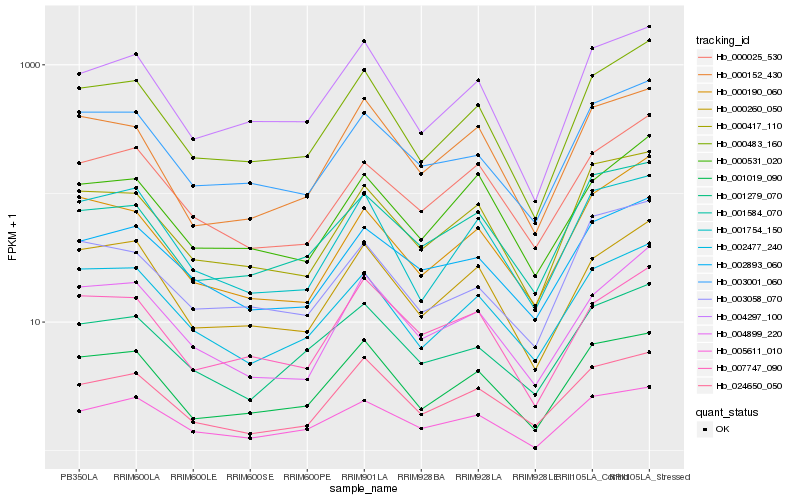
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000483_160 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L34 [Jatropha curcas] |
| 2 |
Hb_004297_100 |
0.056284911 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 3 |
Hb_000260_050 |
0.0692479199 |
- |
- |
PREDICTED: serine/threonine-protein kinase AtPK2/AtPK19-like [Jatropha curcas] |
| 4 |
Hb_001019_090 |
0.0705589144 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000417_110 |
0.0753949411 |
- |
- |
PREDICTED: uncharacterized protein LOC105649681 [Jatropha curcas] |
| 6 |
Hb_002477_240 |
0.0770252247 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 7 |
Hb_004899_220 |
0.077329895 |
- |
- |
PREDICTED: adrenodoxin-like protein 2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_003001_060 |
0.0790336973 |
- |
- |
wound-responsive family protein [Populus trichocarpa] |
| 9 |
Hb_001754_150 |
0.0836952234 |
- |
- |
PREDICTED: vesicle-associated membrane protein 724 [Jatropha curcas] |
| 10 |
Hb_000190_060 |
0.0846273 |
- |
- |
PREDICTED: uncharacterized protein LOC105644688 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000531_020 |
0.0868748026 |
- |
- |
PREDICTED: uncharacterized protein At4g28440-like [Populus euphratica] |
| 12 |
Hb_005611_010 |
0.08789446 |
- |
- |
PREDICTED: protein prune homolog [Jatropha curcas] |
| 13 |
Hb_007747_090 |
0.0897414854 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 14 |
Hb_000025_530 |
0.0906660069 |
- |
- |
PREDICTED: reactive oxygen species modulator 1 [Jatropha curcas] |
| 15 |
Hb_024650_050 |
0.0910535276 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_001584_070 |
0.0921496496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001279_070 |
0.0929905361 |
- |
- |
PREDICTED: uncharacterized protein LOC105633300 [Jatropha curcas] |
| 18 |
Hb_000152_430 |
0.0930774373 |
- |
- |
PREDICTED: 40S ribosomal protein S20-1 [Jatropha curcas] |
| 19 |
Hb_002893_060 |
0.0949853123 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 50 [Jatropha curcas] |
| 20 |
Hb_003058_070 |
0.0957906614 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |