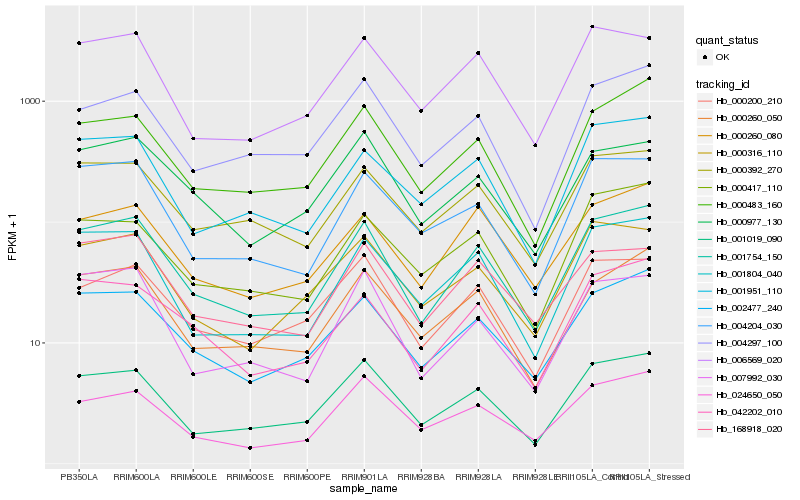
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_150 |
0.0 |
- |
- |
PREDICTED: vesicle-associated membrane protein 724 [Jatropha curcas] |
| 2 |
Hb_002477_240 |
0.0618949944 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 3 |
Hb_001019_090 |
0.0677875326 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001804_040 |
0.0740798885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000260_050 |
0.0758608155 |
- |
- |
PREDICTED: serine/threonine-protein kinase AtPK2/AtPK19-like [Jatropha curcas] |
| 6 |
Hb_042202_010 |
0.079360817 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 7 |
Hb_000392_270 |
0.0827928273 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 8 |
Hb_000483_160 |
0.0836952234 |
- |
- |
PREDICTED: 60S ribosomal protein L34 [Jatropha curcas] |
| 9 |
Hb_004204_030 |
0.0843311844 |
- |
- |
unknown [Populus trichocarpa] |
| 10 |
Hb_000260_080 |
0.0861073384 |
- |
- |
thioredoxin H-type 1 [Hevea brasiliensis] |
| 11 |
Hb_001951_110 |
0.0874036623 |
- |
- |
PREDICTED: DNA-binding protein DDB_G0278111 [Jatropha curcas] |
| 12 |
Hb_024650_050 |
0.0886945704 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000200_210 |
0.0901227599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006569_020 |
0.0910253493 |
- |
- |
hypothetical protein POPTR_0010s22990g [Populus trichocarpa] |
| 15 |
Hb_000316_110 |
0.0922522378 |
- |
- |
Dolichyl-phosphate beta-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_004297_100 |
0.0922601505 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 17 |
Hb_007992_030 |
0.0948176687 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105646951 [Jatropha curcas] |
| 18 |
Hb_000417_110 |
0.0950729267 |
- |
- |
PREDICTED: uncharacterized protein LOC105649681 [Jatropha curcas] |
| 19 |
Hb_168918_020 |
0.0961120024 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 20 |
Hb_000977_130 |
0.0972795428 |
- |
- |
PREDICTED: U6 snRNA-associated Sm-like protein LSm6 [Populus euphratica] |