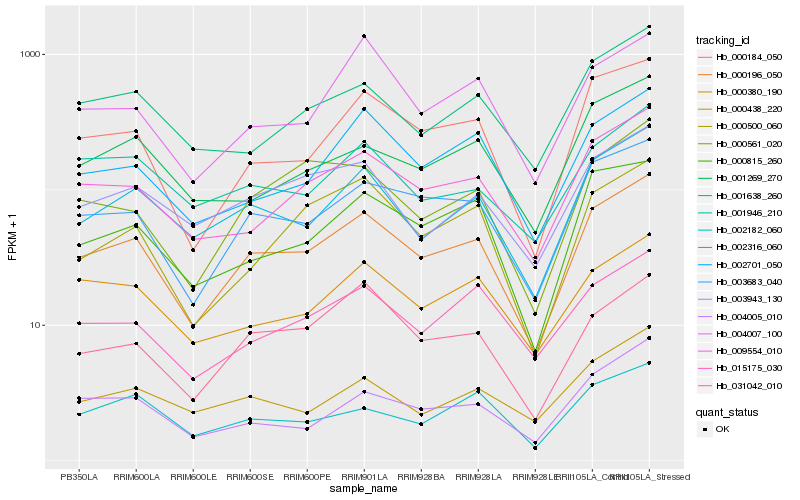
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000196_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 2 |
Hb_003683_040 |
0.0689709822 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_002316_060 |
0.0846088892 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 4 |
Hb_000184_050 |
0.0859401918 |
- |
- |
NOI, putative [Ricinus communis] |
| 5 |
Hb_009554_010 |
0.0878384511 |
- |
- |
PREDICTED: multifunctional methyltransferase subunit TRM112-like protein At1g22270 [Jatropha curcas] |
| 6 |
Hb_000815_260 |
0.0974618097 |
- |
- |
50S ribosomal protein L32pA [Hevea brasiliensis] |
| 7 |
Hb_031042_010 |
0.10091638 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 8 |
Hb_002701_050 |
0.1028849067 |
- |
- |
PREDICTED: 60S ribosomal protein L7a-like [Jatropha curcas] |
| 9 |
Hb_003943_130 |
0.1061737797 |
- |
- |
PREDICTED: uncharacterized protein LOC105636341 [Jatropha curcas] |
| 10 |
Hb_015175_030 |
0.1066207667 |
- |
- |
PREDICTED: RNA-binding protein 24-A [Jatropha curcas] |
| 11 |
Hb_001638_260 |
0.1081608181 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P1 [Vitis vinifera] |
| 12 |
Hb_000561_020 |
0.1081840623 |
- |
- |
hypothetical protein JCGZ_19196 [Jatropha curcas] |
| 13 |
Hb_000380_190 |
0.1101243252 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 14 |
Hb_000500_060 |
0.1112834377 |
- |
- |
BnaC08g21190D [Brassica napus] |
| 15 |
Hb_002182_060 |
0.1128296354 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATXR6 [Jatropha curcas] |
| 16 |
Hb_004005_010 |
0.1149346285 |
- |
- |
PREDICTED: uncharacterized protein LOC102623333 isoform X2 [Citrus sinensis] |
| 17 |
Hb_004007_100 |
0.11540787 |
- |
- |
PREDICTED: 60S ribosomal protein L26-1 [Jatropha curcas] |
| 18 |
Hb_001946_210 |
0.1154149875 |
- |
- |
ribosomal protein l7ae, putative [Ricinus communis] |
| 19 |
Hb_001269_270 |
0.1154602418 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 20 |
Hb_000438_220 |
0.1184108873 |
- |
- |
PREDICTED: uncharacterized protein LOC105649213 [Jatropha curcas] |