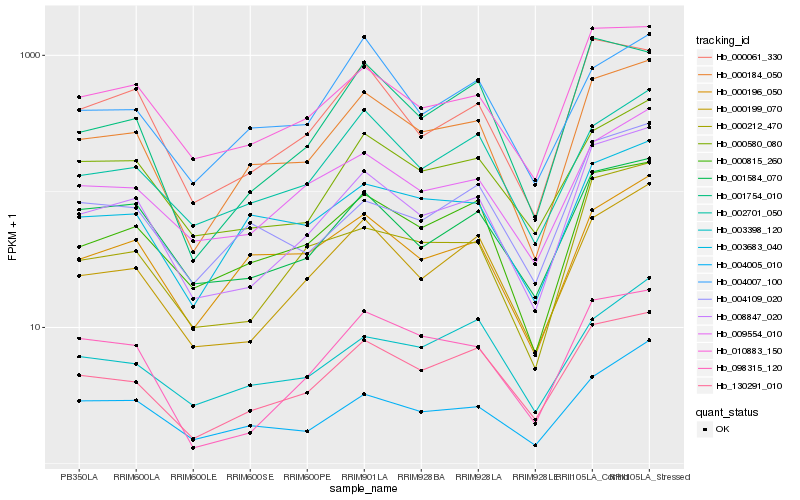
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000184_050 |
0.0 |
- |
- |
NOI, putative [Ricinus communis] |
| 2 |
Hb_130291_010 |
0.0763829676 |
- |
- |
PREDICTED: protein PEROXIN-4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000815_260 |
0.0765530603 |
- |
- |
50S ribosomal protein L32pA [Hevea brasiliensis] |
| 4 |
Hb_003683_040 |
0.077309139 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_008847_020 |
0.0796979561 |
- |
- |
40S ribosomal protein S17C [Hevea brasiliensis] |
| 6 |
Hb_000196_050 |
0.0859401918 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 7 |
Hb_010883_150 |
0.0964011156 |
- |
- |
Ubiquitin-fold modifier 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000199_070 |
0.0979701846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002701_050 |
0.0982721358 |
- |
- |
PREDICTED: 60S ribosomal protein L7a-like [Jatropha curcas] |
| 10 |
Hb_009554_010 |
0.1025944789 |
- |
- |
PREDICTED: multifunctional methyltransferase subunit TRM112-like protein At1g22270 [Jatropha curcas] |
| 11 |
Hb_000580_080 |
0.1044285781 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 12 |
Hb_000061_330 |
0.1116797453 |
- |
- |
eukaryotic translation initiation factor 5A isoform VII [Hevea brasiliensis] |
| 13 |
Hb_001584_070 |
0.1118315367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004005_010 |
0.1131040079 |
- |
- |
PREDICTED: uncharacterized protein LOC102623333 isoform X2 [Citrus sinensis] |
| 15 |
Hb_004007_100 |
0.1133578346 |
- |
- |
PREDICTED: 60S ribosomal protein L26-1 [Jatropha curcas] |
| 16 |
Hb_000212_470 |
0.1179303519 |
- |
- |
hypothetical protein CICLE_v10033256mg [Citrus clementina] |
| 17 |
Hb_098315_120 |
0.1182247492 |
- |
- |
- |
| 18 |
Hb_001754_010 |
0.1192794799 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 19 |
Hb_003398_120 |
0.1197353915 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 20 |
Hb_004109_020 |
0.1215784943 |
- |
- |
60S ribosomal protein L18aA [Hevea brasiliensis] |