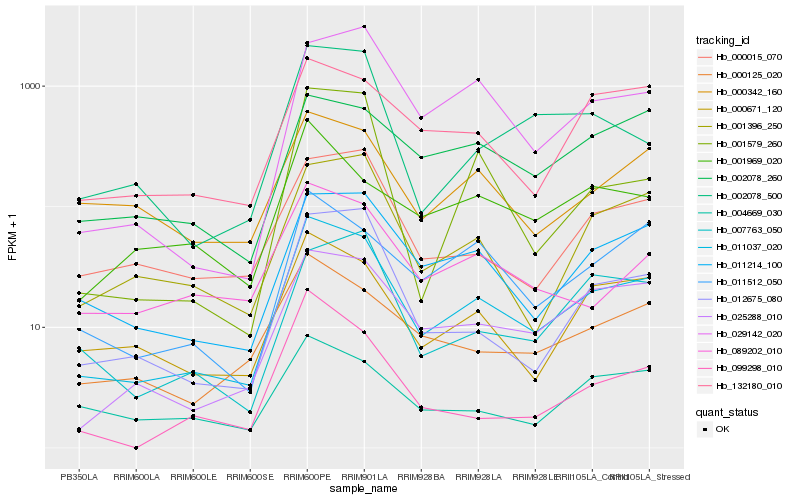
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011037_020 |
0.0 |
- |
- |
unknown [Picea sitchensis] |
| 2 |
Hb_025288_010 |
0.1305001027 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 3 |
Hb_000671_120 |
0.1307484516 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 4 |
Hb_011214_100 |
0.1314012435 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 5 |
Hb_012675_080 |
0.1467757169 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 6 |
Hb_000125_020 |
0.1469710718 |
transcription factor |
TF Family: mTERF |
- |
| 7 |
Hb_011512_050 |
0.1509604222 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 8 |
Hb_001396_250 |
0.1526695898 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 9 |
Hb_132180_010 |
0.1573848884 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 10 |
Hb_000015_070 |
0.1575067271 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 11 |
Hb_089202_010 |
0.162571218 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 12 |
Hb_029142_020 |
0.163535954 |
- |
- |
hypothetical protein POPTR_0004s23900g [Populus trichocarpa] |
| 13 |
Hb_002078_500 |
0.1668654915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004669_030 |
0.1694887063 |
- |
- |
- |
| 15 |
Hb_000342_160 |
0.1698599556 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 16 |
Hb_007763_050 |
0.1712435561 |
- |
- |
hypothetical protein L484_012752 [Morus notabilis] |
| 17 |
Hb_001579_260 |
0.171744672 |
- |
- |
putative ADP-ribolylation factor [Arabidopsis thaliana] |
| 18 |
Hb_002078_260 |
0.1719039066 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_001969_020 |
0.1754284504 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 20 |
Hb_099298_010 |
0.1763873953 |
- |
- |
- |