| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
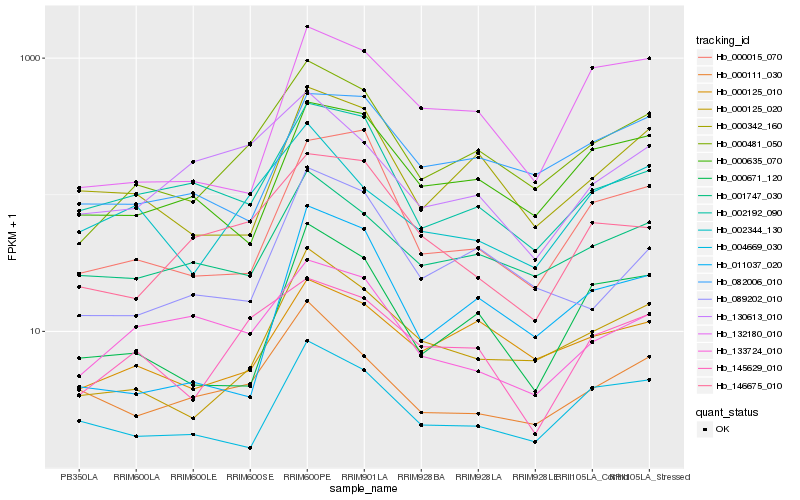
Hb_000481_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636546 [Jatropha curcas] |
| 2 |
Hb_000125_020 |
0.1184623408 |
transcription factor |
TF Family: mTERF |
- |
| 3 |
Hb_000125_010 |
0.1341424268 |
transcription factor |
TF Family: mTERF |
- |
| 4 |
Hb_001747_030 |
0.1361603358 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 5 |
Hb_145629_010 |
0.1411363465 |
- |
- |
- |
| 6 |
Hb_000342_160 |
0.1498781713 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 7 |
Hb_089202_010 |
0.150979298 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 8 |
Hb_000635_070 |
0.1545058623 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 9 |
Hb_002192_090 |
0.1551683055 |
- |
- |
hypothetical protein AMTR_s00029p00055000 [Amborella trichopoda] |
| 10 |
Hb_082006_010 |
0.1572292393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000111_030 |
0.1606839676 |
- |
- |
PREDICTED: uncharacterized protein LOC104094030 [Nicotiana tomentosiformis] |
| 12 |
Hb_000671_120 |
0.1607604173 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 13 |
Hb_002344_130 |
0.1622815505 |
- |
- |
PREDICTED: copper transporter 5 [Jatropha curcas] |
| 14 |
Hb_146675_010 |
0.1630252616 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier-like isoform X2 [Malus domestica] |
| 15 |
Hb_000015_070 |
0.1672245504 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 16 |
Hb_130613_010 |
0.1682344681 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 17 |
Hb_004669_030 |
0.1750847317 |
- |
- |
- |
| 18 |
Hb_132180_010 |
0.1761948717 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 19 |
Hb_133724_010 |
0.1774984748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_011037_020 |
0.1827104409 |
- |
- |
unknown [Picea sitchensis] |