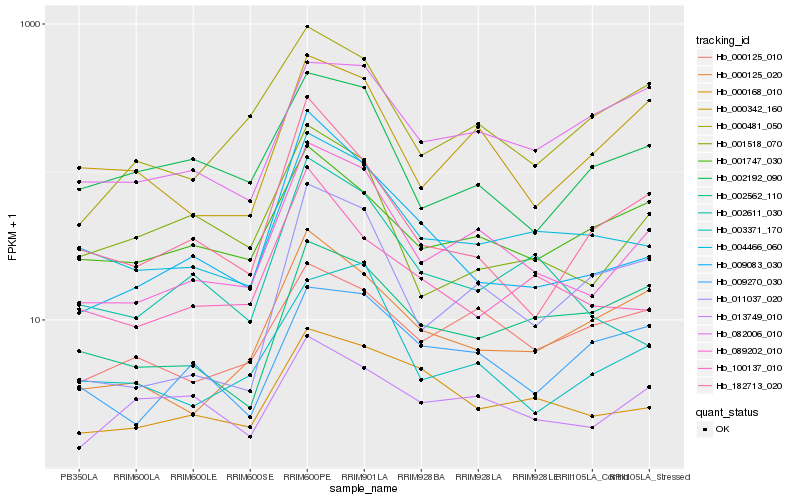
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089202_010 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 2 |
Hb_004466_060 |
0.1317909232 |
- |
- |
hypothetical protein POPTR_0009s12110g [Populus trichocarpa] |
| 3 |
Hb_000342_160 |
0.1448290329 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 4 |
Hb_000481_050 |
0.150979298 |
- |
- |
PREDICTED: uncharacterized protein LOC105636546 [Jatropha curcas] |
| 5 |
Hb_003371_170 |
0.1568734727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000168_010 |
0.1612759912 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3-like isoform X1 [Populus euphratica] |
| 7 |
Hb_000125_020 |
0.1618681253 |
transcription factor |
TF Family: mTERF |
- |
| 8 |
Hb_011037_020 |
0.162571218 |
- |
- |
unknown [Picea sitchensis] |
| 9 |
Hb_002192_090 |
0.1632210115 |
- |
- |
hypothetical protein AMTR_s00029p00055000 [Amborella trichopoda] |
| 10 |
Hb_001518_070 |
0.1635499938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_009083_030 |
0.1636736952 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 12 |
Hb_001747_030 |
0.1663397827 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 13 |
Hb_013749_010 |
0.1674682607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_182713_020 |
0.168494903 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |
| 15 |
Hb_000125_010 |
0.169595734 |
transcription factor |
TF Family: mTERF |
- |
| 16 |
Hb_002562_110 |
0.1753179491 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 17 |
Hb_002611_030 |
0.1767701532 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 18 |
Hb_009270_030 |
0.1789026156 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 19 |
Hb_100137_010 |
0.1798811748 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 20 |
Hb_082006_010 |
0.1803655361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |